DNA Topology and the Initiation of Virus DNA Packaging
- PMID: 27144448
- PMCID: PMC4856287
- DOI: 10.1371/journal.pone.0154785
DNA Topology and the Initiation of Virus DNA Packaging
Abstract
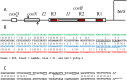
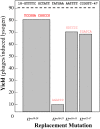
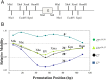
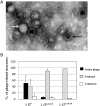
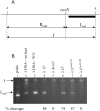
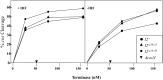
During progeny assembly, viruses selectively package virion genomes from a nucleic acid pool that includes host nucleic acids. For large dsDNA viruses, including tailed bacteriophages and herpesviruses, immature viral DNA is recognized and translocated into a preformed icosahedral shell, the prohead. Recognition involves specific interactions between the viral packaging enzyme, terminase, and viral DNA recognition sites. Generally, viral DNA is recognized by terminase's small subunit (TerS). The large terminase subunit (TerL) contains translocation ATPase and endonuclease domains. In phage lambda, TerS binds a sequence repeated three times in cosB, the recognition site. TerS binding to cosB positions TerL to cut the concatemeric DNA at the adjacent nicking site, cosN. TerL introduces staggered nicks in cosN, generating twelve bp cohesive ends. Terminase separates the cohesive ends and remains bound to the cosB-containing end, in a nucleoprotein structure called Complex I. Complex I docks on the prohead's portal vertex and translocation ensues. DNA topology plays a role in the TerSλ-cosBλ interaction. Here we show that a site, I2, located between cosN and cosB, is critically important for an early DNA packaging step. I2 contains a complex static bend. I2 mutations block DNA packaging. I2 mutant DNA is cut by terminase at cosN in vitro, but in vivo, no cos cleavage is detected, nor is there evidence for Complex I. Models for what packaging step might be blocked by I2 mutations are presented.
Conflict of interest statement
Figures
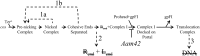
Similar articles
-
Mutations in Nu1, the gene encoding the small subunit of bacteriophage lambda terminase, suppress the postcleavage DNA packaging defect of cosB mutations.J Bacteriol. 1997 Apr;179(8):2479-85. doi: 10.1128/jb.179.8.2479-2485.1997. J Bacteriol. 1997. PMID: 9098042 Free PMC article.
-
Virus DNA packaging: the strategy used by phage lambda.Mol Microbiol. 1995 Jun;16(6):1075-86. doi: 10.1111/j.1365-2958.1995.tb02333.x. Mol Microbiol. 1995. PMID: 8577244 Review.
-
The role of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda, in the nicking reaction.J Mol Biol. 1993 Dec 5;234(3):594-609. doi: 10.1006/jmbi.1993.1614. J Mol Biol. 1993. PMID: 8254662
-
DNA Packaging Specificity of Bacteriophage N15 with an Excursion into the Genetics of a Cohesive End Mismatch.PLoS One. 2015 Dec 3;10(12):e0141934. doi: 10.1371/journal.pone.0141934. eCollection 2015. PLoS One. 2015. PMID: 26633301 Free PMC article.
-
Mechanisms of DNA Packaging by Large Double-Stranded DNA Viruses.Annu Rev Virol. 2015 Nov;2(1):351-78. doi: 10.1146/annurev-virology-100114-055212. Epub 2015 Sep 10. Annu Rev Virol. 2015. PMID: 26958920 Free PMC article. Review.
Cited by
-
Portal Protein: The Orchestrator of Capsid Assembly for the dsDNA Tailed Bacteriophages and Herpesviruses.Annu Rev Virol. 2019 Sep 29;6(1):141-160. doi: 10.1146/annurev-virology-092818-015819. Epub 2019 Jul 23. Annu Rev Virol. 2019. PMID: 31337287 Free PMC article. Review.
-
Viral Genomic DNA Packaging Machinery.Subcell Biochem. 2024;104:181-205. doi: 10.1007/978-3-031-58843-3_9. Subcell Biochem. 2024. PMID: 38963488 Free PMC article. Review.
-
Efficient synthesis of CRISPR-Cas13a-antimicrobial capsids against MRSA facilitated by silent mutation incorporation.Sci Rep. 2024 Jul 13;14(1):16225. doi: 10.1038/s41598-024-67193-5. Sci Rep. 2024. PMID: 39003336 Free PMC article.
References
-
- Catalano CE. Viral genome packaging machines: genetics, structure, and mechanism. Georgetown, Tex.; New York, N.Y: Landes Bioscience/Eurekah.com; Kluwer Academic/Plenum Publishers; 2005. 153 p.
-
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nature reviewsMicrobiology. 2011;9(9):647. - PubMed
-
- Tavares P, Lurz R, Stiege A, Ruckert B, Trautner TA. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. Journal of molecular biology. 1996;264(5):954 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

