Collecting duct carcinoma of the kidney is associated with CDKN2A deletion and SLC family gene up-regulation
- PMID: 27144525
- PMCID: PMC5058651
- DOI: 10.18632/oncotarget.9093
Collecting duct carcinoma of the kidney is associated with CDKN2A deletion and SLC family gene up-regulation
Abstract
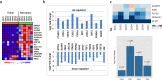
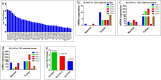
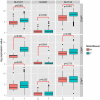
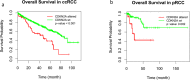
The genetic landscape and molecular features of collecting duct carcinoma (CDC) of the kidney remain largely unknown. Herein, we performed whole exome sequencing (WES) and transcriptome sequencing (RNASeq) on 7 CDC samples (CDC1 -7). Among the 7 samples, 4 samples with matched non-tumor tissue were used for copy number analysis by SNP array data. No recurrent somatic SNVs were observed except for MLL, which was found to be mutated (p.V297I and p.F407C) in 2 samples. We identified somatic SNVs in 14 other cancer census genes including: ATM, CREBBP, PRDM1, CBFB, FBXW7, IKZF1, KDR, KRAS, NACA, NF2, NUP98, SS18, TP53, and ZNF521. SNP array data identified a CDKN2A homozygous deletion in 3 samples and SNV analysis showed a non-sense mutation of the CDKN2A gene with unknown somatic status. To estimate the recurrent rate of CDKN2A abnormalities, we performed FISH screening of additional samples and confirmed the frequent loss (62.5%) of CDKN2A expression. Since cisplatin based therapy is the common treatment option for CDC, we investigated the expression of solute carrier (SLC) family transporters and found 45% alteration. In addition, SLC7A11 (cystine transporter, xCT), a cisplatin resistance associated gene, was found to be overexpressed in 4 out of 5 (80%) cases of CDC tumors tested, as compared to matched non-tumor tissue. In summary, our study provides a comprehensive genomic analysis of CDC and identifies potential pathways suitable for targeted therapies.
Keywords: CDKN2A; collecting duct carcinoma; solute carrier family genes.
Conflict of interest statement
The authors do not have any conflicts of interest.
Figures






References
-
- Gupta R, Billis A, Shah RB, Moch H, Osunkoya AO, Jochum W, Hes O, Bacchi CE, de Castro MG, Hansel DE, Zhou M, Vankalakunti M, Salles PG, Cabrera RA, Gown AM, Amin MB. Carcinoma of the collecting ducts of Bellini and renal medullary carcinoma: clinicopathologic analysis of 52 cases of rare aggressive subtypes of renal cell carcinoma with a focus on their interrelationship. Am J Surg Pathol. 2012;36:1265–1278. - PubMed
-
- Srigley JR, Delahunt B. Uncommon and recently described renal carcinomas. Mod Pathol. 2009;22(Suppl 2):S2–S23. - PubMed
-
- Tokuda N, Naito S, Matsuzaki O, Nagashima Y, Ozono S, Igarashi T, Japanese Society of Renal C. Collecting duct (Bellini duct) renal cell carcinoma: a nationwide survey in Japan. The Journal of urology. 2006;176:40–43. discussion 43. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

