A new automatic algorithm for quantification of myocardial infarction imaged by late gadolinium enhancement cardiovascular magnetic resonance: experimental validation and comparison to expert delineations in multi-center, multi-vendor patient data
- PMID: 27145749
- PMCID: PMC4855857
- DOI: 10.1186/s12968-016-0242-5
A new automatic algorithm for quantification of myocardial infarction imaged by late gadolinium enhancement cardiovascular magnetic resonance: experimental validation and comparison to expert delineations in multi-center, multi-vendor patient data
Abstract
Background: Late gadolinium enhancement (LGE) cardiovascular magnetic resonance (CMR) using magnitude inversion recovery (IR) or phase sensitive inversion recovery (PSIR) has become clinical standard for assessment of myocardial infarction (MI). However, there is no clinical standard for quantification of MI even though multiple methods have been proposed. Simple thresholds have yielded varying results and advanced algorithms have only been validated in single center studies. Therefore, the aim of this study was to develop an automatic algorithm for MI quantification in IR and PSIR LGE images and to validate the new algorithm experimentally and compare it to expert delineations in multi-center, multi-vendor patient data.
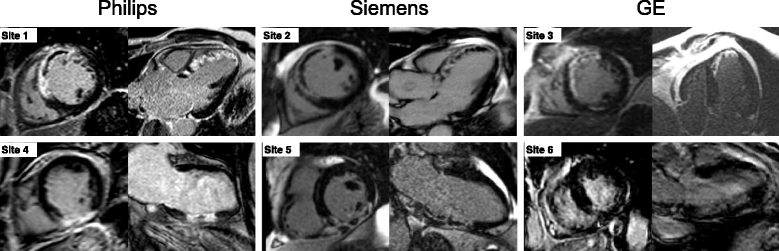
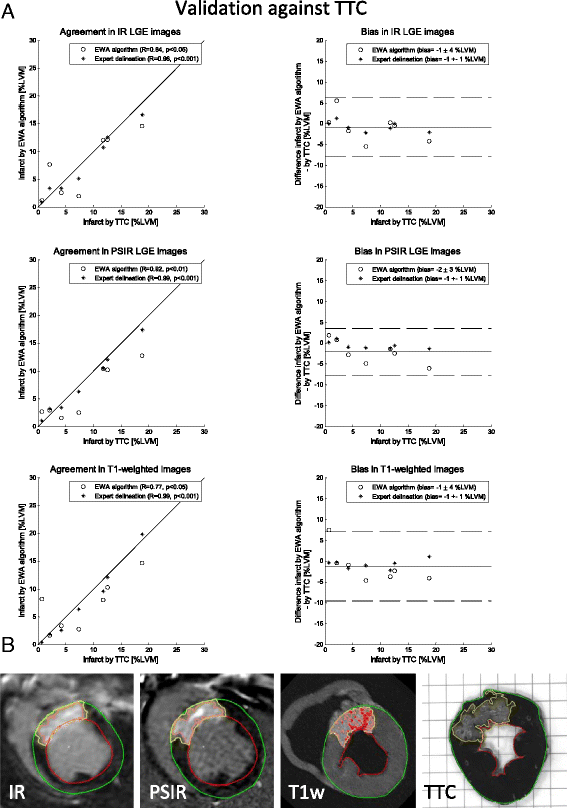
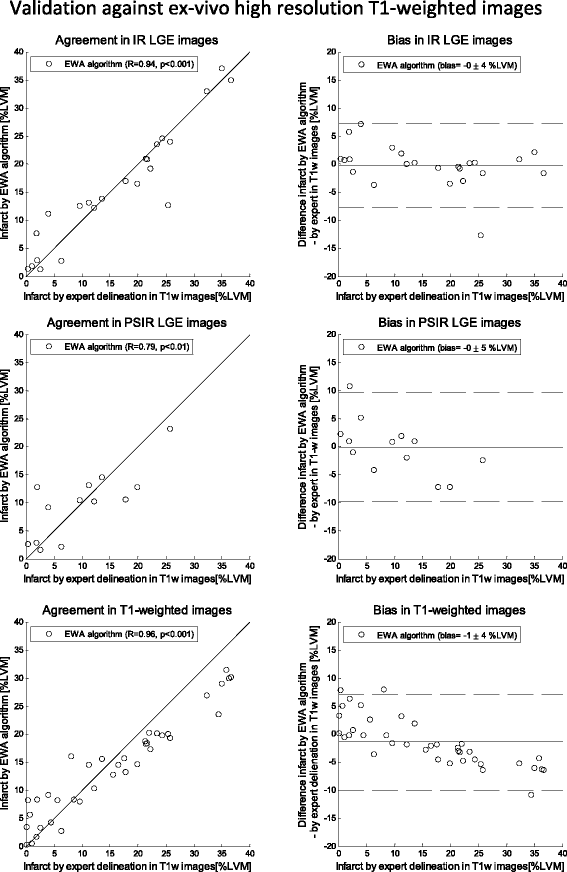
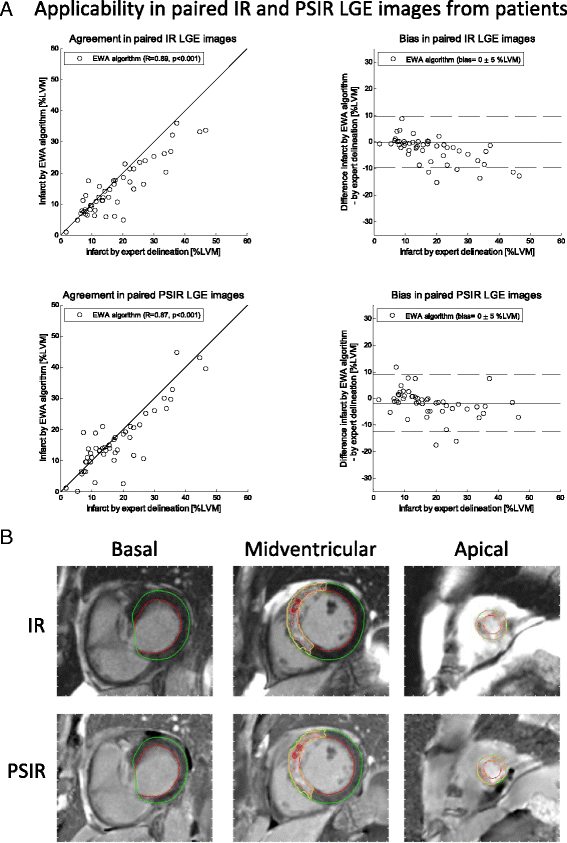
Methods: The new automatic algorithm, EWA (Expectation Maximization, weighted intensity, a priori information), was implemented using an intensity threshold by Expectation Maximization (EM) and a weighted summation to account for partial volume effects. The EWA algorithm was validated in-vivo against triphenyltetrazolium-chloride (TTC) staining (n = 7 pigs with paired IR and PSIR images) and against ex-vivo high resolution T1-weighted images (n = 23 IR and n = 13 PSIR images). The EWA algorithm was also compared to expert delineation in 124 patients from multi-center, multi-vendor clinical trials 2-6 days following first time ST-elevation myocardial infarction (STEMI) treated with percutaneous coronary intervention (PCI) (n = 124 IR and n = 49 PSIR images).
Results: Infarct size by the EWA algorithm in vivo in pigs showed a bias to ex-vivo TTC of -1 ± 4%LVM (R = 0.84) in IR and -2 ± 3%LVM (R = 0.92) in PSIR images and a bias to ex-vivo T1-weighted images of 0 ± 4%LVM (R = 0.94) in IR and 0 ± 5%LVM (R = 0.79) in PSIR images. In multi-center patient studies, infarct size by the EWA algorithm showed a bias to expert delineation of -2 ± 6 %LVM (R = 0.81) in IR images (n = 124) and 0 ± 5%LVM (R = 0.89) in PSIR images (n = 49).
Conclusions: The EWA algorithm was validated experimentally and in patient data with a low bias in both IR and PSIR LGE images. Thus, the use of EM and a weighted intensity as in the EWA algorithm, may serve as a clinical standard for the quantification of myocardial infarction in LGE CMR images.
Clinical trial registration: CHILL-MI: NCT01379261 .
Mitocare: NCT01374321 .
Keywords: Automatic quantification algorithm; Expectation maximization; Experimental validation; LGE CMR; Multi-center patient data.
Figures

References
-
- Fieno DS, Kim RJ, Chen EL, Lomasney JW, Klocke FJ, Judd RM. Contrast-enhanced magnetic resonance imaging of myocardium at risk: distinction between reversible and irreversible injury throughout infarct healing. J Am Coll Cardiol. 2000;36(6):1985–91. doi: 10.1016/S0735-1097(00)00958-X. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

