New Perspectives on Genomic Imprinting, an Essential and Multifaceted Mode of Epigenetic Control in the Developing and Adult Brain
- PMID: 27145912
- PMCID: PMC5125552
- DOI: 10.1146/annurev-neuro-061010-113708
New Perspectives on Genomic Imprinting, an Essential and Multifaceted Mode of Epigenetic Control in the Developing and Adult Brain
Abstract
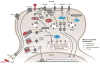
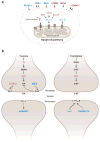
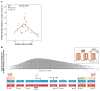
Mammalian evolution entailed multiple innovations in gene regulation, including the emergence of genomic imprinting, an epigenetic regulation leading to the preferential expression of a gene from its maternal or paternal allele. Genomic imprinting is highly prevalent in the brain, yet, until recently, its central roles in neural processes have not been fully appreciated. Here, we provide a comprehensive survey of adult and developmental brain functions influenced by imprinted genes, from neural development and wiring to synaptic function and plasticity, energy balance, social behaviors, emotions, and cognition. We further review the widespread identification of parental biases alongside monoallelic expression in brain tissues, discuss their potential roles in dosage regulation of key neural pathways, and suggest possible mechanisms underlying the dynamic regulation of imprinting in the brain. This review should help provide a better understanding of the significance of genomic imprinting in the normal and pathological brain of mammals including humans.
Keywords: behavior; energy balance; gene dosage; genomic imprinting; neural development; parental bias.
Figures



References
-
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17(1):75–78. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

