RNase E-based degradosome modulates polyadenylation of mRNAs after Rho-independent transcription terminators in Escherichia coli
- PMID: 27145979
- PMCID: PMC5149407
- DOI: 10.1111/mmi.13413
RNase E-based degradosome modulates polyadenylation of mRNAs after Rho-independent transcription terminators in Escherichia coli
Abstract
Here we demonstrate that the RNase E-based degradosome is required for poly(A) polymerase I (PAP I)-dependent polyadenylation after Rho-independent transcription terminators for both mono- and polycistronic transcripts. Disruption of degradosome assembly in mutants lacking the polynucleotide phosphorylase (PNPase) binding domain led to a significant increase in the level of PNPase synthesized polynucleotide tails in the rpsJ and rpsM polycistronic transcripts and the lpp monocistronic transcript. The polynucleotide tails were mostly located within the coding sequences in the degradosome mutants compared to the wild type control where the majority of the PAP I synthesized poly(A) tails were after the Rho-independent transcription terminators. For the Rho terminated metNIQ operon, the tails for all three mRNAs were predominately polynucleotide and were located within the coding sequences in both wild type and degradosome mutant strains. Furthermore, by employing a pnp-R100D point mutant that encodes a catalytically inactive PNPase protein that still forms intact degradosomes, we show that a catalytically active PNPase is required for normal mRNA polyadenylation by PAP I. Our data suggest that polyadenylation requires a functional degradosome to maintain an equilibrium between free PNPase and the PAP I polyadenylation complex.
© 2016 John Wiley & Sons Ltd.
Figures
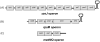
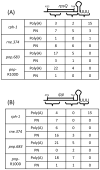
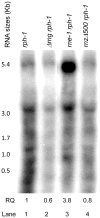
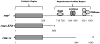
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

