Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals
- PMID: 27146893
- PMCID: PMC4884081
- DOI: 10.7554/eLife.15690
Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals
Abstract
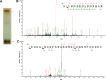
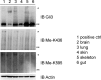
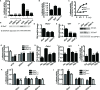
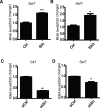
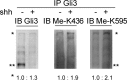
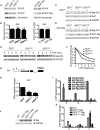
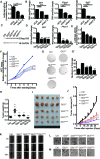
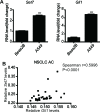
Hedgehog signaling plays very important roles in development and cancers. Vertebrates have three transcriptional factors, Gli1, Gli2 and Gli3. Among them, Gli3 is a very special transcriptional factor which closely resembles Cubitus interruptus (Ci, in Drosophila) structurally and functionally as a 'double agent' for Shh target gene expression. Here we show that Gli3 full-length, but not the truncated form, can be methylated at K436 and K595. This methylation is specifically catalyzed by Set7, a lysine methyltransferase (KMT). Methylation at K436 and K595 respectively increases the stability and DNA binding ability of Gli3, resulting in an enhancement of Shh signaling activation. Furthermore, functional experiments indicate that the Gli3 methylation contributes to the tumor growth and metastasis in non-small cell lung cancer in vitro and in vivo. Therefore, we propose that Set7 mediated methylation is a novel PTM of Gli3, which positively regulates the transactivity of Gli3 and the activation of Shh signaling.
Keywords: Gli3; Set7; biochemistry; cancer biology; hedgehog signaling; methylation; mouse; tumorigenesis.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



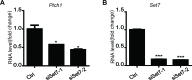
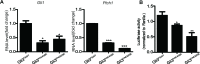

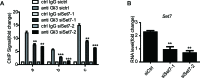
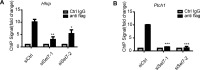

Similar articles
-
Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3.J Biol Chem. 1999 Mar 19;274(12):8143-52. doi: 10.1074/jbc.274.12.8143. J Biol Chem. 1999. PMID: 10075717
-
GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis.Development. 2006 Feb;133(3):569-78. doi: 10.1242/dev.02220. Epub 2006 Jan 5. Development. 2006. PMID: 16396903
-
PKA-mediated Gli2 and Gli3 phosphorylation is inhibited by Hedgehog signaling in cilia and reduced in Talpid3 mutant.Dev Biol. 2017 Sep 1;429(1):147-157. doi: 10.1016/j.ydbio.2017.06.035. Epub 2017 Jul 1. Dev Biol. 2017. PMID: 28673820 Free PMC article.
-
Restore the brake on tumor progression.Biochem Pharmacol. 2017 Aug 15;138:1-6. doi: 10.1016/j.bcp.2017.04.003. Epub 2017 Apr 5. Biochem Pharmacol. 2017. PMID: 28389227 Free PMC article. Review.
-
A translational approach to congenital non-obstructive hydronephrosis.Pediatr Nephrol. 2013 Sep;28(9):1757-61. doi: 10.1007/s00467-012-2321-7. Epub 2012 Oct 5. Pediatr Nephrol. 2013. PMID: 23052658 Review.
Cited by
-
Multiprotein GLI Transcriptional Complexes as Therapeutic Targets in Cancer.Life (Basel). 2022 Nov 24;12(12):1967. doi: 10.3390/life12121967. Life (Basel). 2022. PMID: 36556332 Free PMC article. Review.
-
H3 histone methylation landscape in male urogenital cancers: from molecular mechanisms to epigenetic biomarkers and therapeutic targets.Front Cell Dev Biol. 2023 May 9;11:1181764. doi: 10.3389/fcell.2023.1181764. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37228649 Free PMC article. Review.
-
SETD7 Promotes Cell Proliferation and Migration via Methylation-mediated TAF7 in Clear Cell Renal Cell Carcinoma.Int J Biol Sci. 2024 May 19;20(8):3008-3027. doi: 10.7150/ijbs.93201. eCollection 2024. Int J Biol Sci. 2024. PMID: 38904013 Free PMC article.
-
Lysine and arginine methylation of transcription factors.Cell Mol Life Sci. 2024 Dec 16;82(1):5. doi: 10.1007/s00018-024-05531-6. Cell Mol Life Sci. 2024. PMID: 39680066 Free PMC article. Review.
-
Methylated DNMT1 and E2F1 are targeted for proteolysis by L3MBTL3 and CRL4DCAF5 ubiquitin ligase.Nat Commun. 2018 Apr 24;9(1):1641. doi: 10.1038/s41467-018-04019-9. Nat Commun. 2018. PMID: 29691401 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

