Immunomediated Pan-cancer Regulation Networks are Dominant Fingerprints After Treatment of Cell Lines with Demethylation
- PMID: 27147816
- PMCID: PMC4849425
- DOI: 10.4137/CIN.S31809
Immunomediated Pan-cancer Regulation Networks are Dominant Fingerprints After Treatment of Cell Lines with Demethylation
Abstract
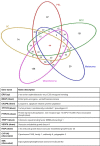
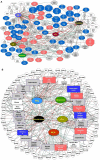
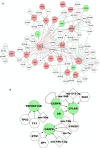
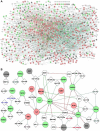
Pan-cancer studies are particularly relevant not only for addressing the complexity of the inherently observed heterogeneity but also for identifying clinically relevant features that may be common to the cancer types. Immune system regulations usually reveal synergistic modulation with other cancer mechanisms and in combination provide insights on possible advances in cancer immunotherapies. Network inference is a powerful approach to decipher pan-cancer systems dynamics. The methodology proposed in this study elucidates the impacts of epigenetic treatment on the drivers of complex pan-cancer regulation circuits involving cell lines of five cancer types. These patterns were observed from differential gene expression measurements following demethylation with 5-azacytidine. Networks were built to establish associations of phenotypes at molecular level with cancer hallmarks through both transcriptional and post-transcriptional regulation mechanisms. The most prominent feature that emerges from our integrative network maps, linking pathway landscapes to disease and drug-target associations, refers primarily to a mosaic of immune-system crosslinked influences. Therefore, characteristics initially evidenced in single cancer maps become motifs well summarized by network cores and fingerprints.
Keywords: demethylation; drug-target maps; gene networks; immunomediated regulation; pan-cancer.
Figures



Similar articles
-
Network assessment of demethylation treatment in melanoma: Differential transcriptome-methylome and antigen profile signatures.PLoS One. 2018 Nov 28;13(11):e0206686. doi: 10.1371/journal.pone.0206686. eCollection 2018. PLoS One. 2018. PMID: 30485296 Free PMC article.
-
Immuno-Oncology Integrative Networks: Elucidating the Influences of Osteosarcoma Phenotypes.Cancer Inform. 2017 Jul 26;16:1176935117721691. doi: 10.1177/1176935117721691. eCollection 2017. Cancer Inform. 2017. PMID: 28804242 Free PMC article. Review.
-
Survey and comparative assessments of computational multi-omics integrative methods with multiple regulatory networks identifying distinct tumor compositions across pan-cancer data sets.Brief Bioinform. 2021 May 20;22(3):bbaa102. doi: 10.1093/bib/bbaa102. Brief Bioinform. 2021. PMID: 32533167
-
Construction of pan-cancer regulatory networks based on causal inference.Biosystems. 2024 Sep;243:105279. doi: 10.1016/j.biosystems.2024.105279. Epub 2024 Jul 23. Biosystems. 2024. PMID: 39053644
-
Time-course gene profiling and networks in demethylated retinoblastoma cell line.Oncotarget. 2015 Sep 15;6(27):23688-707. doi: 10.18632/oncotarget.4644. Oncotarget. 2015. PMID: 26143641 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

