The Role of Complement in Cnidarian-Dinoflagellate Symbiosis and Immune Challenge in the Sea Anemone Aiptasia pallida
- PMID: 27148208
- PMCID: PMC4840205
- DOI: 10.3389/fmicb.2016.00519
The Role of Complement in Cnidarian-Dinoflagellate Symbiosis and Immune Challenge in the Sea Anemone Aiptasia pallida
Abstract
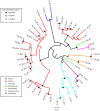
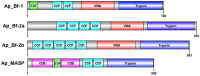
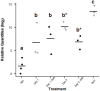
The complement system is an innate immune pathway that in vertebrates, is responsible for initial recognition and ultimately phagocytosis and destruction of microbes. Several complement molecules including C3, Factor B, and mannose binding lectin associated serine proteases (MASP) have been characterized in invertebrates and while most studies have focused on their conserved role in defense against pathogens, little is known about their role in managing beneficial microbes. The purpose of this study was to (1) characterize complement pathway genes in the symbiotic sea anemone Aiptasia pallida, (2) investigate the evolution of complement genes in invertebrates, and (3) examine the potential dual role of complement genes Factor B and MASP in the onset and maintenance of cnidarian-dinoflagellate symbiosis and immune challenge using qPCR based studies. The results demonstrate that A. pallida has multiple Factor B genes (Ap_Bf-1, Ap_Bf-2a, and Ap_Bf-2b) and one MASP gene (Ap_MASP). Phylogenetic analysis indicates that the evolutionary history of complement genes is complex, and there have been many gene duplications or gene loss events, even within members of the same phylum. Gene expression analyses revealed a potential role for complement in both onset and maintenance of cnidarian-dinoflagellate symbiosis and immune challenge. Specifically, Ap_Bf-1 and Ap_MASP are significantly upregulated in the light at the onset of symbiosis and in response to challenge with the pathogen Serratia marcescens suggesting that they play a role in the initial recognition of both beneficial and harmful microbes. Ap_Bf-2b in contrast, was generally downregulated during the onset and maintenance of symbiosis and in response to challenge with S. marcescens. Therefore, the exact role of Ap_Bf-2b in response to microbes remains unclear, but the results suggest that the presence of microbes leads to repressed expression. Together, these results indicate functional divergence between Ap_Bf-1 and Ap_Bf-2b, and that Ap_Bf-1 and Ap_MASP may be functioning together in an ancestral hybrid of the lectin and alternative complement pathways. Overall, this study provides information on the role of the complement system in a basal metazoan and its role in host-microbe interactions.
Keywords: Aiptasia; Serratia marcescens; Symbiodinium; cnidarians; complement; innate immunity; symbiosis.
Figures






References
-
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

