The Whole Genome Assembly and Comparative Genomic Research of Thellungiella parvula (Extremophile Crucifer) Mitochondrion
- PMID: 27148547
- PMCID: PMC4842374
- DOI: 10.1155/2016/5283628
The Whole Genome Assembly and Comparative Genomic Research of Thellungiella parvula (Extremophile Crucifer) Mitochondrion
Abstract
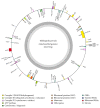
The complete nucleotide sequences of the mitochondrial (mt) genome of an extremophile species Thellungiella parvula (T. parvula) have been determined with the lengths of 255,773 bp. T. parvula mt genome is a circular sequence and contains 32 protein-coding genes, 19 tRNA genes, and three ribosomal RNA genes with a 11.5% coding sequence. The base composition of 27.5% A, 27.5% T, 22.7% C, and 22.3% G in descending order shows a slight bias of 55% AT. Fifty-three repeats were identified in the mitochondrial genome of T. parvula, including 24 direct repeats, 28 tandem repeats (TRs), and one palindromic repeat. Furthermore, a total of 199 perfect microsatellites have been mined with a high A/T content (83.1%) through simple sequence repeat (SSR) analysis and they were distributed unevenly within this mitochondrial genome. We also analyzed other plant mitochondrial genomes' evolution in general, providing clues for the understanding of the evolution of organelles genomes in plants. Comparing with other Brassicaceae species, T. parvula is related to Arabidopsis thaliana whose characters of low temperature resistance have been well documented. This study will provide important genetic tools for other Brassicaceae species research and improve yields of economically important plants.
Figures



Similar articles
-
Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis.Plant Physiol. 2010 Nov;154(3):1040-52. doi: 10.1104/pp.110.163923. Epub 2010 Sep 10. Plant Physiol. 2010. PMID: 20833729 Free PMC article.
-
The genome of the extremophile crucifer Thellungiella parvula.Nat Genet. 2011 Aug 7;43(9):913-8. doi: 10.1038/ng.889. Nat Genet. 2011. PMID: 21822265 Free PMC article.
-
The whole genome assembly and evolution analyze of carmine radish (Raphanus sativus L.) Mitochondrion.Mitochondrial DNA B Resour. 2020 Jun 1;5(3):2252-2253. doi: 10.1080/23802359.2020.1772136. eCollection 2020. Mitochondrial DNA B Resour. 2020. PMID: 33366995 Free PMC article.
-
Syntenic gene analysis between Brassica rapa and other Brassicaceae species.Front Plant Sci. 2012 Aug 30;3:198. doi: 10.3389/fpls.2012.00198. eCollection 2012. Front Plant Sci. 2012. PMID: 22969786 Free PMC article.
-
Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis-a case of "Tandem duplication-random loss" for genome rearrangement in Crassostrea?BMC Genomics. 2008 Oct 11;9:477. doi: 10.1186/1471-2164-9-477. BMC Genomics. 2008. PMID: 18847502 Free PMC article.
Cited by
-
Comparative analyses of three complete Primula mitogenomes with insights into mitogenome size variation in Ericales.BMC Genomics. 2022 Nov 24;23(1):770. doi: 10.1186/s12864-022-08983-x. BMC Genomics. 2022. PMID: 36424546 Free PMC article.
-
Halophytism: What Have We Learnt From Arabidopsis thaliana Relative Model Systems?Plant Physiol. 2018 Nov;178(3):972-988. doi: 10.1104/pp.18.00863. Epub 2018 Sep 20. Plant Physiol. 2018. PMID: 30237204 Free PMC article. Review.
-
Organellar genome assembly methods and comparative analysis of horticultural plants.Hortic Res. 2018 Jan 10;5:3. doi: 10.1038/s41438-017-0002-1. eCollection 2018. Hortic Res. 2018. PMID: 29423233 Free PMC article.
-
Characterization of the complete chloroplast genome of Camellia brevistyla, an oil-rich and evergreen shrub.Mitochondrial DNA B Resour. 2020 Jan 21;5(1):386-387. doi: 10.1080/23802359.2019.1703607. Mitochondrial DNA B Resour. 2020. PMID: 33366568 Free PMC article.
References
-
- Tanaka Y., Tsuda M., Yasumoto K., Yamagishi H., Terachi T. A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.) BMC Genomics. 2012;13(1, article 352) doi: 10.1186/1471-2164-13-352. - DOI - PMC - PubMed
-
- Abdelnoor R. V., Yule R., Elo A., Christensen A. C., Meyer-Gauen G., Mackenzie S. A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5968–5973. doi: 10.1073/pnas.1037651100. - DOI - PMC - PubMed
-
- Alverson A. J., Wei X., Rice D. W., Stern D. B., Barry K., Palmer J. D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Molecular Biology and Evolution. 2010;27(6):1436–1448. doi: 10.1093/molbev/msq029. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

