Complete hematologic response of early T-cell progenitor acute lymphoblastic leukemia to the γ-secretase inhibitor BMS-906024: genetic and epigenetic findings in an outlier case
- PMID: 27148573
- PMCID: PMC4850884
- DOI: 10.1101/mcs.a000539
Complete hematologic response of early T-cell progenitor acute lymphoblastic leukemia to the γ-secretase inhibitor BMS-906024: genetic and epigenetic findings in an outlier case
Abstract
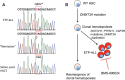
Notch pathway antagonists such as γ-secretase inhibitors (GSIs) are being tested in diverse cancers, but exceptional responses have yet to be reported. We describe the case of a patient with relapsed/refractory early T-cell progenitor acute lymphoblastic leukemia (ETP-ALL) who achieved a complete hematologic response following treatment with the GSI BMS-906024. Whole-exome sequencing of leukemic blasts revealed heterozygous gain-of-function driver mutations in NOTCH1, CSF3R, and PTPN11, and a homozygous/hemizygous loss-of-function mutation in DNMT3A. The three gain-of-function mutations were absent from remission marrow cells, but the DNMT3A mutation persisted in heterozygous form in remission marrow, consistent with an origin for the patient's ETP-ALL from clonal hematopoiesis. Ex vivo culture of ETP-ALL blasts confirmed high levels of activated NOTCH1 that were repressed by GSI treatment, and RNA-seq documented that GSIs downregulated multiple known Notch target genes. Surprisingly, one potential target gene that was unaffected by GSIs was MYC, a key Notch target in GSI-sensitive T-ALL of cortical T-cell type. H3K27ac super-enhancer landscapes near MYC showed a pattern previously reported in acute myeloid leukemia (AML) that is sensitive to BRD4 inhibitors, and in line with this ETP-ALL blasts downregulated MYC in response to the BRD4 inhibitor JQ1. To our knowledge, this is the first example of complete response of a Notch-mutated ETP-ALL to a Notch antagonist and is also the first description of chromatin landscapes associated with ETP-ALL. Our experience suggests that additional attempts to target Notch in Notch-mutated ETP-ALL are merited.
Keywords: leukemia.
Figures




References
-
- Aster JC, Blacklow SC. 2012. Targeting the Notch pathway: twists and turns on the road to rational therapeutics. J Clin Oncol 30: 2418–2420. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
