Chromatoid Body Protein TDRD6 Supports Long 3' UTR Triggered Nonsense Mediated mRNA Decay
- PMID: 27149095
- PMCID: PMC4858158
- DOI: 10.1371/journal.pgen.1005857
Chromatoid Body Protein TDRD6 Supports Long 3' UTR Triggered Nonsense Mediated mRNA Decay
Abstract
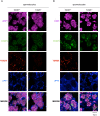
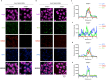
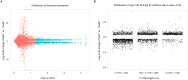
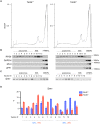
Chromatoid bodies (CBs) are spermiogenesis-specific organelles of largely unknown function. CBs harbor various RNA species, RNA-associated proteins and proteins of the tudor domain family like TDRD6, which is required for a proper CB architecture. Proteome analysis of purified CBs revealed components of the nonsense-mediated mRNA decay (NMD) machinery including UPF1. TDRD6 is essential for UPF1 localization to CBs, for UPF1-UPF2 and UPF1-MVH interactions. Upon removal of TDRD6, the association of several mRNAs with UPF1 and UPF2 is disturbed, and the long 3' UTR-stimulated but not the downstream exon-exon junction triggered pathway of NMD is impaired. Reduced association of the long 3' UTR mRNAs with UPF1 and UPF2 correlates with increased stability and enhanced translational activity. Thus, we identified TDRD6 within CBs as required for mRNA degradation, specifically the extended 3' UTR-triggered NMD pathway, and provide evidence for the requirement of NMD in spermiogenesis. This function depends on TDRD6-promoted assembly of mRNA and decay enzymes in CBs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Comment in
-
Spermatogenesis Studies Reveal a Distinct Nonsense-Mediated mRNA Decay (NMD) Mechanism for mRNAs with Long 3'UTRs.PLoS Genet. 2016 May 5;12(5):e1005979. doi: 10.1371/journal.pgen.1005979. eCollection 2016 May. PLoS Genet. 2016. PMID: 27149371 Free PMC article. No abstract available.
References
-
- Chalmel F, Lardenois A, Primig M (2007) Toward understanding the core meiotic transcriptome in mammals and its implications for somatic cancer. Ann N Y Acad Sci 1120: 1–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

