UPF2-Dependent Nonsense-Mediated mRNA Decay Pathway Is Essential for Spermatogenesis by Selectively Eliminating Longer 3'UTR Transcripts
- PMID: 27149259
- PMCID: PMC4858225
- DOI: 10.1371/journal.pgen.1005863
UPF2-Dependent Nonsense-Mediated mRNA Decay Pathway Is Essential for Spermatogenesis by Selectively Eliminating Longer 3'UTR Transcripts
Abstract
During transcription, most eukaryotic genes generate multiple alternative cleavage and polyadenylation (APA) sites, leading to the production of transcript isoforms with variable lengths in the 3' untranslated region (3'UTR). In contrast to somatic cells, male germ cells, especially pachytene spermatocytes and round spermatids, express a distinct reservoir of mRNAs with shorter 3'UTRs that are essential for spermatogenesis and male fertility. However, the mechanisms underlying the enrichment of shorter 3'UTR transcripts in the developing male germ cells remain unknown. Here, we report that UPF2-mediated nonsense-mediated mRNA decay (NMD) plays an essential role in male germ cells by eliminating ubiquitous genes-derived, longer 3'UTR transcripts, and that this role is independent of its canonical role in degrading "premature termination codon" (PTC)-containing transcripts in somatic cell lineages. This report provides physiological evidence supporting a noncanonical role of the NMD pathway in achieving global 3'UTR shortening in the male germ cells during spermatogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
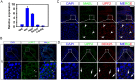
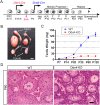
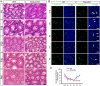
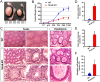



Comment in
-
Spermatogenesis Studies Reveal a Distinct Nonsense-Mediated mRNA Decay (NMD) Mechanism for mRNAs with Long 3'UTRs.PLoS Genet. 2016 May 5;12(5):e1005979. doi: 10.1371/journal.pgen.1005979. eCollection 2016 May. PLoS Genet. 2016. PMID: 27149371 Free PMC article. No abstract available.
References
-
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiological reviews. 1972;52(1):198–236. Epub 1972/01/01. . - PubMed
-
- Eddy EM. Male germ cell gene expression. Recent progress in hormone research. 2002;57:103–28. Epub 2002/05/23. . - PubMed
-
- Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenetic and genome research. 2003;103(3–4):217–24. Epub 2004/03/31. 76807. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

