RalGPS2 Is Essential for Survival and Cell Cycle Progression of Lung Cancer Cells Independently of Its Established Substrates Ral GTPases
- PMID: 27149377
- PMCID: PMC4858283
- DOI: 10.1371/journal.pone.0154840
RalGPS2 Is Essential for Survival and Cell Cycle Progression of Lung Cancer Cells Independently of Its Established Substrates Ral GTPases
Abstract
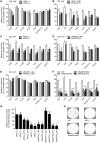
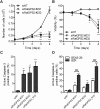
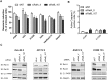
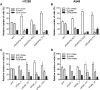
The human genome contains six genes coding for proteins validated in vitro as specific activators of the small GTPases "Ras-related protein Ral-A" and "Ras-related protein Ral-B", generically named Ral-guanine nucleotide exchange factors (RalGEF). Ral proteins are important contributors to Ras oncogenic signaling, and RAS oncogenes are important in human Non-Small Cell Lung Carcinoma (NSCLC). Therefore in this work, RalGEF contribution to oncogenic and non-oncogenic features of human NSCLC cell lines, as anchorage-dependent and independent growth, cell survival, and proliferation, was investigated. Among all human RalGEF, silencing of RGL1 and RALGPS1 had no detectable effect. However, silencing of either RGL2, RGL3, RALGDS or, to a larger extent, RALGPS2 inhibited cell population growth in anchorage dependent and independent conditions (up to 90 and 80%, respectively). RALGPS2 silencing also caused an increase in the number of apoptotic cells, up to 45% of the cell population in transformed bronchial BZR cells. In H1299 and A549, two NSCLC cell lines, RALGPS2 silencing caused an arrest of cells in the G0/G1-phase of cell cycle. Furthermore, it was associated with the modulation of important cell cycle regulators: the E3 Ubiquitin Protein Ligase S-phase kinase-associated protein 2 (Skp2) was strongly down-regulated (both at mRNA and protein levels), and its targets, the cell cycle inhibitors p27 and p21, were up-regulated. These molecular effects were not mimicked by silencing RALA, RALB, or both. However, RALB silencing caused a modest inhibition of cell cycle progression, which in H1299 cells was associated with Cyclin D1 regulation. In conclusion, RALGPS2 is implicated in the control of cell cycle progression and survival in the in vitro growth of NSCLC cell lines. This function is largely independent of Ral GTPases and associated with modulation of Skp2, p27 and p21 cell cycle regulators.
Conflict of interest statement
Figures


Similar articles
-
Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms.J Biol Chem. 2010 Nov 5;285(45):34729-40. doi: 10.1074/jbc.M110.116756. Epub 2010 Aug 27. J Biol Chem. 2010. PMID: 20801877 Free PMC article.
-
RalGPS2 is involved in tunneling nanotubes formation in 5637 bladder cancer cells.Exp Cell Res. 2018 Jan 15;362(2):349-361. doi: 10.1016/j.yexcr.2017.11.036. Epub 2017 Dec 6. Exp Cell Res. 2018. PMID: 29208460
-
Silencing the FOLR2 Gene Inhibits Cell Proliferation and Increases Apoptosis in the NCI-H1650 Non-Small Cell Lung Cancer Cell Line via Inhibition of AKT/Mammalian Target of Rapamycin (mTOR)/Ribosomal Protein S6 Kinase 1 (S6K1) Signaling.Med Sci Monit. 2018 Nov 11;24:8064-8073. doi: 10.12659/MSM.911384. Med Sci Monit. 2018. PMID: 30415267 Free PMC article.
-
RalGDS family members couple Ras to Ral signalling and that's not all.Cell Signal. 2010 Dec;22(12):1804-10. doi: 10.1016/j.cellsig.2010.05.010. Epub 2010 May 15. Cell Signal. 2010. PMID: 20478380 Review.
-
The RAL signaling network: Cancer and beyond.Int Rev Cell Mol Biol. 2021;361:21-105. doi: 10.1016/bs.ircmb.2020.10.005. Epub 2020 Dec 2. Int Rev Cell Mol Biol. 2021. PMID: 34074494 Review.
Cited by
-
RGL2 Drives the Metastatic Progression of Colorectal Cancer via Preventing the Protein Degradation of β-Catenin and KRAS.Cancers (Basel). 2021 Apr 7;13(8):1763. doi: 10.3390/cancers13081763. Cancers (Basel). 2021. PMID: 33917100 Free PMC article.
-
Transcriptomic signatures of cellular and humoral immune responses in older adults after seasonal influenza vaccination identified by data-driven clustering.Sci Rep. 2018 Jan 15;8(1):739. doi: 10.1038/s41598-017-17735-x. Sci Rep. 2018. PMID: 29335477 Free PMC article.
-
A general calculus of fitness landscapes finds genes under selection in cancers.Genome Res. 2022 May;32(5):916-929. doi: 10.1101/gr.275811.121. Epub 2022 Mar 17. Genome Res. 2022. PMID: 35301263 Free PMC article.
-
Integrative Analysis of Somatic Mutations in Non-coding Regions Altering RNA Secondary Structures in Cancer Genomes.Sci Rep. 2019 Jun 3;9(1):8205. doi: 10.1038/s41598-019-44489-5. Sci Rep. 2019. PMID: 31160636 Free PMC article.
-
Identification of HIF-2α-regulated genes that play a role in human microvascular endothelial sprouting during prolonged hypoxia in vitro.Angiogenesis. 2017 Feb;20(1):39-54. doi: 10.1007/s10456-016-9527-4. Epub 2016 Oct 3. Angiogenesis. 2017. PMID: 27699500 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

