Distinct role of Arabidopsis mitochondrial P-type pentatricopeptide repeat protein-modulating editing protein, PPME, in nad1 RNA editing
- PMID: 27149614
- PMCID: PMC4962808
- DOI: 10.1080/15476286.2016.1184384
Distinct role of Arabidopsis mitochondrial P-type pentatricopeptide repeat protein-modulating editing protein, PPME, in nad1 RNA editing
Abstract
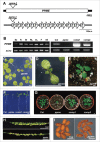
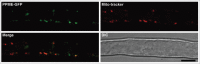
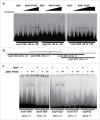
The mitochondrion is an important power generator in most eukaryotic cells. To preserve its function, many essential nuclear-encoded factors play specific roles in mitochondrial RNA metabolic processes, including RNA editing. RNA editing consists of post-transcriptional deamination, which alters specific nucleotides in transcripts to mediate gene expression. In plant cells, many pentatricopeptide repeat proteins (PPRs) participate in diverse organellar RNA metabolic processes, but only PLS-type PPRs are involved in RNA editing. Here, we report a P-type PPR protein from Arabidopsis thaliana, P-type PPR-Modulating Editing (PPME), which has a distinct role in mitochondrial nad1 RNA editing via RNA binding activity. In the homozygous ppme mutant, cytosine (C)-to-uracil (U) conversions at both the nad1-898 and 937 sites were abolished, disrupting Arg(300)-to-Trp(300) and Pro(313)-to-Ser(313) amino acid changes in the mitochondrial NAD1 protein. NAD1 is a critical component of mitochondrial respiration complex I; its activity is severely reduced in the homozygous ppme mutant, resulting in significantly altered growth and development. Both abolished RNA editing and defective complex I activity were completely rescued by CaMV 35S promoter- and PPME native promoter-driven PPME genomic fragments tagged with GFP in a homozygous ppme background. Our experimental results demonstrate a distinct role of a P-type PPR protein, PPME, in RNA editing in plant organelles.
Keywords: Arabidopsis; PPR; complex I activity; editing; mitochondria; nad1.
Figures


References
-
- Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UCM, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998; 396:133-40; PMID:9823893; http://dx.doi.org/10.1038/24094 - DOI - PubMed
-
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science 1999; 283: 1476-81; PMID:10066161; http://dx.doi.org/10.1126/science.283.5407.1476 - DOI - PubMed
-
- Burger G, Gray MW, Franz Lang B. Mitochondrial genomes: anything goes. Trends in Genetics 2003; 19: 709-16; PMID:14642752; http://dx.doi.org/10.1016/j.tig.2003.10.012 - DOI - PubMed
-
- Murcha MW, Wang Y, Narsai R, Whelan J. The plant mitochondrial protein import apparatus - the differences make it interesting. Biochim Biophys Acta 2014; 1840: 1233-45; PMID:24080405; http://dx.doi.org/10.1016/j.bbagen.2013.09.026 - DOI - PubMed
-
- Liere K, Weihe A, Borner T. The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 2011; 168: 1345-60; PMID:21316793; http://dx.doi.org/10.1016/j.jplph.2011.01.005 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
