PRDM16 Suppresses MLL1r Leukemia via Intrinsic Histone Methyltransferase Activity
- PMID: 27151440
- PMCID: PMC5061593
- DOI: 10.1016/j.molcel.2016.03.010
PRDM16 Suppresses MLL1r Leukemia via Intrinsic Histone Methyltransferase Activity
Abstract
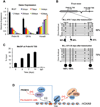
PRDM16 is a transcription co-factor that plays critical roles in development of brown adipose tissue, as well as maintenance of adult hematopoietic and neural stem cells. Here we report that PRDM16 is a histone H3K4 methyltransferase on chromatin. Mutation in the N-terminal PR domain of PRDM16 abolishes the intrinsic enzymatic activity of PRDM16. We show that the methyltransferase activity of PRDM16 is required for specific suppression of MLL fusion protein-induced leukemogenesis both in vitro and in vivo. Mechanistic studies show that PRDM16 directly activates the SNAG family transcription factor Gfi1b, which in turn downregulates the HOXA gene cluster. Knockdown Gfi1b represses PRDM16-mediated tumor suppression, while Gfi1b overexpression mimics PRDM16 overexpression. In further support of the tumor suppressor function of PRDM16, silencing PRDM16 by DNA methylation is concomitant with MLL-AF9-induced leukemic transformation. Taken together, our study reveals a previously uncharacterized function of PRDM16 that depends on its PR domain activity.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

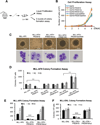
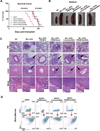
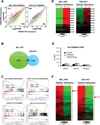

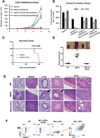
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

