Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants
- PMID: 27152936
- PMCID: PMC5148187
- DOI: 10.1038/ismej.2016.62
Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants
Abstract
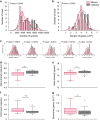
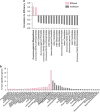
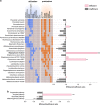
Ruminants have the remarkable ability to convert human-indigestible plant biomass into human-digestible food products, due to a complex microbiome residing in the rumen compartment of their upper digestive tract. Here we report the discovery that rumen microbiome components are tightly linked to cows' ability to extract energy from their feed, termed feed efficiency. Feed efficiency was measured in 146 milking cows and analyses of the taxonomic composition, gene content, microbial activity and metabolomic composition was performed on the rumen microbiomes from the 78 most extreme animals. Lower richness of microbiome gene content and taxa was tightly linked to higher feed efficiency. Microbiome genes and species accurately predicted the animals' feed efficiency phenotype. Specific enrichment of microbes and metabolic pathways in each of these microbiome groups resulted in better energy and carbon channeling to the animal, while lowering methane emissions to the atmosphere. This ecological and mechanistic understanding of the rumen microbiome could lead to an increase in available food resources and environmentally friendly livestock agriculture.
Figures




References
-
- Aha DW. (1997) Lazy Learning. Kluwer Academic Publishers: Norwell, MA, USA.
-
- Ajmone-Marsan P, Garcia JF, Lenstra JA. (2010). On the origin of cattle: how aurochs became cattle and colonized the world. Evol Anthropol Issues News Rev 19: 148–157.
-
- Archer JA, Richardson EC, Herd RM, Arthur PF. (1999). Potential for selection to improve efficiency of feed use in beef cattle. Aust J Agric Res 50: 147–162.
-
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 57: 289–300.
-
- Bradford GE. (1999). Contributions of animal agriculture to meeting global human food demand. Livest Prod Sci 59: 95–112.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

