Dynamics of Translation of Single mRNA Molecules In Vivo
- PMID: 27153498
- PMCID: PMC4889334
- DOI: 10.1016/j.cell.2016.04.034
Dynamics of Translation of Single mRNA Molecules In Vivo
Abstract
Regulation of mRNA translation, the process by which ribosomes decode mRNAs into polypeptides, is used to tune cellular protein levels. Currently, methods for observing the complete process of translation from single mRNAs in vivo are unavailable. Here, we report the long-term (>1 hr) imaging of single mRNAs undergoing hundreds of rounds of translation in live cells, enabling quantitative measurements of ribosome initiation, elongation, and stalling. This approach reveals a surprising heterogeneity in the translation of individual mRNAs within the same cell, including rapid and reversible transitions between a translating and non-translating state. Applying this method to the cell-cycle gene Emi1, we find strong overall repression of translation initiation by specific 5' UTR sequences, but individual mRNA molecules in the same cell can exhibit dramatically different translational efficiencies. The ability to observe translation of single mRNA molecules in live cells provides a powerful tool to study translation regulation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

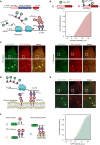










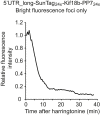
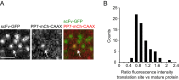

Comment in
-
Translation: Live stream: translation at single-mRNA resolution.Nat Rev Genet. 2016 Jun 16;17(7):373. doi: 10.1038/nrg.2016.79. Nat Rev Genet. 2016. PMID: 27306872 No abstract available.
-
PROTEIN TRANSLATION. Seeing translation.Science. 2016 Jun 17;352(6292):1391-2. doi: 10.1126/science.aag1039. Science. 2016. PMID: 27313023 No abstract available.
-
Tracking translation one mRNA at a time.Nat Biotechnol. 2016 Jul 12;34(7):723-4. doi: 10.1038/nbt.3632. Nat Biotechnol. 2016. PMID: 27404883 No abstract available.
References
-
- Aakalu G., Smith W.B., Nguyen N., Jiang C., Schuman E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. - PubMed
-
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

