Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells
- PMID: 27153499
- PMCID: PMC4905760
- DOI: 10.1016/j.cell.2016.04.040
Real-Time Imaging of Translation on Single mRNA Transcripts in Live Cells
Abstract
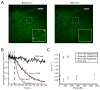
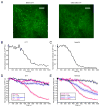
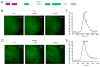
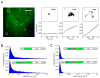
Translation is under tight spatial and temporal controls to ensure protein production in the right time and place in cells. Methods that allow real-time, high-resolution visualization of translation in live cells are essential for understanding the spatiotemporal dynamics of translation regulation. Based on multivalent fluorescence amplification of the nascent polypeptide signal, we develop a method to image translation on individual mRNA molecules in real time in live cells, allowing direct visualization of translation events at the translation sites. Using this approach, we monitor transient changes of translation dynamics in responses to environmental stresses, capture distinct mobilities of individual polysomes in different subcellular compartments, and detect 3' UTR-dependent local translation and active transport of polysomes in dendrites of primary neurons.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Translation: Live stream: translation at single-mRNA resolution.Nat Rev Genet. 2016 Jun 16;17(7):373. doi: 10.1038/nrg.2016.79. Nat Rev Genet. 2016. PMID: 27306872 No abstract available.
-
PROTEIN TRANSLATION. Seeing translation.Science. 2016 Jun 17;352(6292):1391-2. doi: 10.1126/science.aag1039. Science. 2016. PMID: 27313023 No abstract available.
-
Tracking translation one mRNA at a time.Nat Biotechnol. 2016 Jul 12;34(7):723-4. doi: 10.1038/nbt.3632. Nat Biotechnol. 2016. PMID: 27404883 No abstract available.
References
-
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. - PubMed
-
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK) Journal of medicinal chemistry. 2012;55:7193–7207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

