Integrated genome browser: visual analytics platform for genomics
- PMID: 27153568
- PMCID: PMC4937187
- DOI: 10.1093/bioinformatics/btw069
Integrated genome browser: visual analytics platform for genomics
Abstract
Motivation: Genome browsers that support fast navigation through vast datasets and provide interactive visual analytics functions can help scientists achieve deeper insight into biological systems. Toward this end, we developed Integrated Genome Browser (IGB), a highly configurable, interactive and fast open source desktop genome browser.
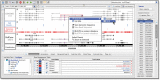
Results: Here we describe multiple updates to IGB, including all-new capabilities to display and interact with data from high-throughput sequencing experiments. To demonstrate, we describe example visualizations and analyses of datasets from RNA-Seq, ChIP-Seq and bisulfite sequencing experiments. Understanding results from genome-scale experiments requires viewing the data in the context of reference genome annotations and other related datasets. To facilitate this, we enhanced IGB's ability to consume data from diverse sources, including Galaxy, Distributed Annotation and IGB-specific Quickload servers. To support future visualization needs as new genome-scale assays enter wide use, we transformed the IGB codebase into a modular, extensible platform for developers to create and deploy all-new visualizations of genomic data.
Availability and implementation: IGB is open source and is freely available from http://bioviz.org/igb
Contact: aloraine@uncc.edu.
© The Author 2016. Published by Oxford University Press.
Figures



References
-
- Bederson B.B, Boltman A. (1999) Does animation help users build mental maps of spatial information? In: Information Visualization. (Info Vis '99) Proceedings. IEEE Symposium, pp. 28–35.
-
- Cockburn A. et al. (2009) A review of overview+detail, zooming, and focus+context interfaces. ACM Comput. Surv., 41, 1–31.
-
- Durbin R, Thierry-Mieg J. (1991) A C. elegans database. In: Documentation, code and data available from anonymous FTP servers at lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk and ncbi.nlm.nih.gov.
-
- Fielding R.T., Taylor R.N. (2002) Principled design of the modern Web architecture. ACM Trans. Internet Technol., 2, 115–150.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

