Confidence assignment for mass spectrometry based peptide identifications via the extreme value distribution
- PMID: 27153659
- PMCID: PMC5939896
- DOI: 10.1093/bioinformatics/btw225
Confidence assignment for mass spectrometry based peptide identifications via the extreme value distribution
Abstract
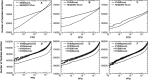
Motivation: There is a growing trend for biomedical researchers to extract evidence and draw conclusions from mass spectrometry based proteomics experiments, the cornerstone of which is peptide identification. Inaccurate assignments of peptide identification confidence thus may have far-reaching and adverse consequences. Although some peptide identification methods report accurate statistics, they have been limited to certain types of scoring function. The extreme value statistics based method, while more general in the scoring functions it allows, demands accurate parameter estimates and requires, at least in its original design, excessive computational resources. Improving the parameter estimate accuracy and reducing the computational cost for this method has two advantages: it provides another feasible route to accurate significance assessment, and it could provide reliable statistics for scoring functions yet to be developed.
Results: We have formulated and implemented an efficient algorithm for calculating the extreme value statistics for peptide identification applicable to various scoring functions, bypassing the need for searching large random databases.
Availability and implementation: The source code, implemented in C ++ on a linux system, is available for download at ftp://ftp.ncbi.nlm.nih.gov/pub/qmbp/qmbp_ms/RAId/RAId_Linux_64Bit
Contact: yyu@ncbi.nlm.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
Published by Oxford University Press 2016. This work is written by US Government employees and is in the public domain in the US.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

