UniProt-DAAC: domain architecture alignment and classification, a new method for automatic functional annotation in UniProtKB
- PMID: 27153729
- PMCID: PMC4965628
- DOI: 10.1093/bioinformatics/btw114
UniProt-DAAC: domain architecture alignment and classification, a new method for automatic functional annotation in UniProtKB
Abstract
Motivation: Similarity-based methods have been widely used in order to infer the properties of genes and gene products containing little or no experimental annotation. New approaches that overcome the limitations of methods that rely solely upon sequence similarity are attracting increased attention. One of these novel approaches is to use the organization of the structural domains in proteins.
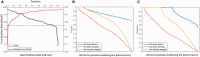
Results: We propose a method for the automatic annotation of protein sequences in the UniProt Knowledgebase (UniProtKB) by comparing their domain architectures, classifying proteins based on the similarities and propagating functional annotation. The performance of this method was measured through a cross-validation analysis using the Gene Ontology (GO) annotation of a sub-set of UniProtKB/Swiss-Prot. The results demonstrate the effectiveness of this approach in detecting functional similarity with an average F-score: 0.85. We applied the method on nearly 55.3 million uncharacterized proteins in UniProtKB/TrEMBL resulted in 44 818 178 GO term predictions for 12 172 114 proteins. 22% of these predictions were for 2 812 016 previously non-annotated protein entries indicating the significance of the value added by this approach.
Availability and implementation: The results of the method are available at: ftp://ftp.ebi.ac.uk/pub/contrib/martin/DAAC/ CONTACT: tdogan@ebi.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures


References
-
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
-
- Bashton M., Chothia C. (2007) The generation of new protein functions by the combination of domains. Structure, 15, 85–99 - PubMed
-
- Björklund ÅK. et al. (2005) Domain rearrangements in protein evolution. J. Mol. Biol., 353, 911–923. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

