ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia
- PMID: 27154821
- PMCID: PMC4930721
- DOI: 10.1158/2159-8290.CD-16-0058
ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia
Abstract
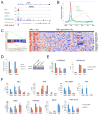
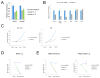
Numerous studies in multiple systems support that histone H3 lysine 36 dimethylation (H3K36me2) is associated with transcriptional activation; however, the underlying mechanisms are not well defined. Here, we show that the H3K36me2 chromatin mark written by the ASH1L histone methyltransferase is preferentially bound in vivo by LEDGF, a mixed-lineage leukemia (MLL)-associated protein that colocalizes with MLL, ASH1L, and H3K36me2 on chromatin genome wide. Furthermore, ASH1L facilitates recruitment of LEDGF and wild-type MLL proteins to chromatin at key leukemia target genes and is a crucial regulator of MLL-dependent transcription and leukemic transformation. Conversely, KDM2A, an H3K36me2 demethylase and Polycomb group silencing protein, antagonizes MLL-associated leukemogenesis. Our studies are the first to provide a basic mechanistic insight into epigenetic interactions wherein placement, interpretation, and removal of H3K36me2 contribute to the regulation of gene expression and MLL leukemia, and suggest ASH1L as a novel target for therapeutic intervention.
Significance: Epigenetic regulators play vital roles in cancer pathogenesis and represent a new frontier in therapeutic targeting. Our studies provide basic mechanistic insight into the role of H3K36me2 in transcription activation and MLL leukemia pathogenesis and implicate ASH1L histone methyltransferase as a promising target for novel molecular therapy. Cancer Discov; 6(7); 770-83. ©2016 AACR.See related commentary by Balbach and Orkin, p. 700This article is highlighted in the In This Issue feature, p. 681.
©2016 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest: The authors have no conflicts of interest to declare.
Figures



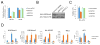

Comment in
-
An Achilles' Heel for MLL-Rearranged Leukemias: Writers and Readers of H3 Lysine 36 Dimethylation.Cancer Discov. 2016 Jul;6(7):700-2. doi: 10.1158/2159-8290.CD-16-0564. Cancer Discov. 2016. PMID: 27371576
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

