Computational identification of genes modulating stem height-diameter allometry
- PMID: 27155207
- PMCID: PMC5103235
- DOI: 10.1111/pbi.12579
Computational identification of genes modulating stem height-diameter allometry
Abstract
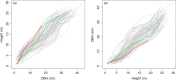
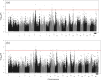
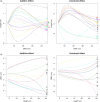
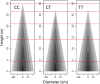
The developmental variation in stem height with respect to stem diameter is related to a broad range of ecological and evolutionary phenomena in trees, but the underlying genetic basis of this variation remains elusive. We implement a dynamic statistical model, functional mapping, to formulate a general procedure for the computational identification of quantitative trait loci (QTLs) that control stem height-diameter allometry during development. Functional mapping integrates the biological principles underlying trait formation and development into the association analysis of DNA genotype and endpoint phenotype, thus providing an incentive for understanding the mechanistic interplay between genes and development. Built on the basic tenet of functional mapping, we explore two core ecological scenarios of how stem height and stem diameter covary in response to environmental stimuli: (i) trees pioneer sunlit space by allocating more growth to stem height than diameter and (ii) trees maintain their competitive advantage through an inverse pattern. The model is equipped to characterize 'pioneering' QTLs (piQTLs) and 'maintaining' QTLs (miQTLs) which modulate these two ecological scenarios, respectively. In a practical application to a mapping population of full-sib hybrids derived from two Populus species, the model has well proven its versatility by identifying several piQTLs that promote height growth at a cost of diameter growth and several miQTLs that benefit radial growth at a cost of height growth. Judicious application of functional mapping may lead to improved strategies for studying the genetic control of the formation mechanisms underlying trade-offs among quantities of assimilates allocated to different growth parts.
Keywords: functional mapping; height-diameter allometry; mathematical equation; quantitative trait loci.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures
Similar articles
-
A Computational Model for Inferring QTL Control Networks Underlying Developmental Covariation.Front Plant Sci. 2019 Dec 18;10:1557. doi: 10.3389/fpls.2019.01557. eCollection 2019. Front Plant Sci. 2019. PMID: 31921232 Free PMC article.
-
Time-specific and pleiotropic quantitative trait loci coordinately modulate stem growth in Populus.Plant Biotechnol J. 2019 Mar;17(3):608-624. doi: 10.1111/pbi.13002. Epub 2018 Sep 17. Plant Biotechnol J. 2019. PMID: 30133117 Free PMC article.
-
A dissection model for mapping complex traits.Plant J. 2019 Mar;97(6):1168-1182. doi: 10.1111/tpj.14185. Epub 2019 Feb 12. Plant J. 2019. PMID: 30536697
-
Integrating Evolutionary Game Theory into Mechanistic Genotype-Phenotype Mapping.Trends Genet. 2016 May;32(5):256-268. doi: 10.1016/j.tig.2016.02.004. Epub 2016 Mar 24. Trends Genet. 2016. PMID: 27017185 Review.
-
Functional mapping of developmental processes: theory, applications, and prospects.Methods Mol Biol. 2012;871:227-43. doi: 10.1007/978-1-61779-785-9_12. Methods Mol Biol. 2012. PMID: 22565840 Review.
Cited by
-
Functional Mapping of Genes Modulating Plant Shade Avoidance Using Leaf Traits.Plants (Basel). 2023 Jan 30;12(3):608. doi: 10.3390/plants12030608. Plants (Basel). 2023. PMID: 36771692 Free PMC article.
-
A Multilayer Interactome Network Constructed in a Forest Poplar Population Mediates the Pleiotropic Control of Complex Traits.Front Genet. 2021 Nov 12;12:769688. doi: 10.3389/fgene.2021.769688. eCollection 2021. Front Genet. 2021. PMID: 34868256 Free PMC article.
-
Genetic dissection of growth trajectories in forest trees: From FunMap to FunGraph.For Res (Fayettev). 2021 Nov 3;1:19. doi: 10.48130/FR-2021-0019. eCollection 2021. For Res (Fayettev). 2021. PMID: 39524511 Free PMC article. Review.
-
A Computational Model for Inferring QTL Control Networks Underlying Developmental Covariation.Front Plant Sci. 2019 Dec 18;10:1557. doi: 10.3389/fpls.2019.01557. eCollection 2019. Front Plant Sci. 2019. PMID: 31921232 Free PMC article.
-
Natural variation in the prolyl 4-hydroxylase gene PtoP4H9 contributes to perennial stem growth in Populus.Plant Cell. 2023 Oct 30;35(11):4046-4065. doi: 10.1093/plcell/koad212. Plant Cell. 2023. PMID: 37522322 Free PMC article.
References
-
- Andersson, B. , Elfving, B. , Persson, T. , Ericsson, T. and Kroon, J. (2007) Characteristics and development of improved Pinus sylvestris in northern Sweden. Can. J. Forest. Res. 37, 84–92.
-
- Bullock, S.H. (2000) Developmental patterns of tree dimensions in a neotropical deciduous forest. Biotropica, 32, 42–52.
-
- Falster, D.S. and Westoby, M. (2003) Plant height and evolutionary games. Trends Ecol. Evol. 18, 337–343.
-
- Feldpausch, T.R. , Banin, L. , Phillips, O.L. , Baker, T.R. , Lewis, S.L. , Quesada, C.A. , Affum‐Baffoe, K. et al. (2011) Height‐diameter allometry of tropical forest trees. Biogeosciences, 8, 1081–1106.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

