Quality control of chemically damaged RNA
- PMID: 27155660
- PMCID: PMC5025279
- DOI: 10.1007/s00018-016-2261-7
Quality control of chemically damaged RNA
Abstract
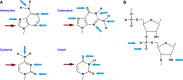
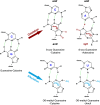
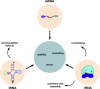
The "central dogma" of molecular biology describes how information contained in DNA is transformed into RNA and finally into proteins. In order for proteins to maintain their functionality in both the parent cell and subsequent generations, it is essential that the information encoded in DNA and RNA remains unaltered. DNA and RNA are constantly exposed to damaging agents, which can modify nucleic acids and change the information they encode. While much is known about how cells respond to damaged DNA, the importance of protecting RNA has only become appreciated over the past decade. Modification of the nucleobase through oxidation and alkylation has long been known to affect its base-pairing properties during DNA replication. Similarly, recent studies have begun to highlight some of the unwanted consequences of chemical damage on mRNA decoding during translation. Oxidation and alkylation of mRNA appear to have drastic effects on the speed and fidelity of protein synthesis. As some mRNAs can persist for days in certain tissues, it is not surprising that it has recently emerged that mRNA-surveillance and RNA-repair pathways have evolved to clear or correct damaged mRNA.
Keywords: 8-Oxoguanosine; Alkylation; O6-Methylguanosine; Oxidation; RNA damage; RNA surveillance; Ribosome; Translation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

