Profiling of the dynamically alteredgene expression in peripheral nerve injury using NGS RNA sequencing technique
- PMID: 27158375
- PMCID: PMC4846932
Profiling of the dynamically alteredgene expression in peripheral nerve injury using NGS RNA sequencing technique
Abstract
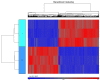
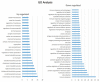
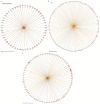
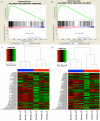
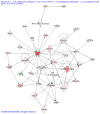
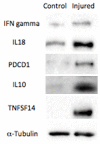
Functional recovery of peripheral nerve injuries is of major demand in clinical practice worldwide. Although, to some extent, peripheral nervous system can spontaneously regenerate, post-injury recovery is often associated with poor functional outcome. The molecular mechanism controlling the peripheral nerve repair process is still majorly unclear. In this study, by utilizing the Next Generation Sequencing (NGS) RNA sequencing technique, we aim to profile the gene expression spectrum of the peripheral nerve repair. In total, we detected 2847 were differentially expressed at day 7 post crush nerve injury. The GO, Panther, IPA and GSEA analysis was performed to decipher the biological processes involving the differentially expressed genes. Collectively, our results highlighted the inflammatory response and related signaling pathway (NFkB and TNFa signaling) play key role in peripheral nerve repair regulation. Furthermore, Network analysis illustrated that the IL10, IL18, IFN-γ and PDCD1 were four key regulators with multiple participations in peripheral nerve repair and potentially exert influence to the repair process. The expression changes of IL10, IL18, IFN-γ, PDCD1 and TNFSF14 (LIGHT) were further validated by western blot analysis. Hopefully, the present study may provide useful platform to further reveal the molecular mechanism of peripheral nerve repair and discover promising treatment target to enhance peripheral nerve regeneration.
Keywords: Peripheral nerve repair; RNA sequencing; inflammation and NFkB.
Figures

References
-
- Sheth S, Li X, Binder S, Dry SM. Differential gene expression profiles of neurothekeomas and nerve sheath myxomas by microarray analysis. Mod Pathol. 2011;24:343–354. - PubMed
-
- Webber C, Zochodne D. The nerve regenerative microenvironment: early behavior and partnership of axons and Schwann cells. Exp Neurol. 2010;223:51–59. - PubMed
-
- Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. - PMC - PubMed
-
- Bosse F, Hasenpusch-Theil K, Kury P, Muller HW. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J Neurochem. 2006;96:1441–1457. - PubMed
-
- Choi YK, Kim KW. Blood-neural barrier: its diversity and coordinated cell-to-cell communication. BMB Rep. 2008;41:345–352. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
