Chronic lymphocytic leukemia cells diversify and differentiate in vivo via a nonclassical Th1-dependent, Bcl-6-deficient process
- PMID: 27158669
- PMCID: PMC4855875
- DOI: 10.1172/jci.insight.86288
Chronic lymphocytic leukemia cells diversify and differentiate in vivo via a nonclassical Th1-dependent, Bcl-6-deficient process
Abstract
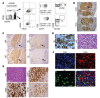
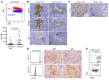
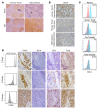
Xenografting primary tumor cells allows modeling of the heterogeneous natures of malignant diseases and the influences of the tissue microenvironment. Here, we demonstrate that xenografting primary chronic lymphocytic leukemia (CLL) B lymphocytes with activated autologous T cells into alymphoid mice results in considerable CLL B cell division and sizable T cell expansion. Nevertheless, most/all CD5+CD19+ cells are eventually lost, due in part to differentiation into antibody-secreting plasmablasts/plasma cells. CLL B cell differentiation is associated with isotype class switching and development of new IGHV-D-J mutations and occurs via an activation-induced deaminase-dependent pathway that upregulates IRF4 and Blimp-1 without appreciable levels of the expected Bcl-6. These processes were induced in IGHV-unmutated and IGHV-mutated clones by Th1-polarized T-bet+ T cells, not classical T follicular helper (Tfh) cells. Thus, the block in B cell maturation, defects in T cell action, and absence of antigen-receptor diversification, which are often cardinal characteristics of CLL, are not inherent but imposed by external signals and the microenvironment. Although these activities are not dominant features in human CLL, each occurs in tissue proliferation centers where the mechanisms responsible for clonal evolution operate. Thus, in this setting, CLL B cell diversification and differentiation develop by a nonclassical germinal center-like reaction that might reflect the cell of origin of this leukemia.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

