Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment
- PMID: 27158675
- PMCID: PMC4855700
- DOI: 10.1172/jci.insight.86355
Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment
Abstract
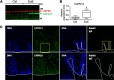
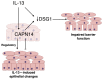
We recently identified a genome-wide genetic association of eosinophilic esophagitis (EoE) at 2p23 spanning the calpain 14 (CAPN14) gene, yet the causal mechanism has not been elucidated. We now show that recombinant CAPN14 cleaves a calpain-specific substrate and is inhibited by 4 classical calpain inhibitors: MDL-28170, acetyl-calpastatin, E-64, and PD151746. CAPN14 is specifically induced (>100-fold) in esophageal epithelium after IL-13 treatment. Epithelial cells overexpressing CAPN14 display impaired epithelial architecture, characterized by acantholysis, epidermal clefting, and epidermolysis. CAPN14 overexpression impairs epithelial barrier function, as demonstrated by decreased transepithelial resistance (2.1-fold) and increased FITC-dextran flux (2.6-fold). Epithelium with gene-silenced CAPN14 demonstrates increased dilated intercellular spaces (5.5-fold) and less organized basal cell layering (1.5-fold) following IL-13 treatment. Finally, CAPN14 overexpression results in loss of desmoglein 1 (DSG1) expression, whereas the IL-13-induced loss of DSG1 is normalized by CAPN14 gene silencing. Importantly, these findings were specific to CAPN14, as they were not observed with modulation of CAPN1 expression. These results, along with the potent induction of CAPN14 by IL-13 and genetic linkage of EoE to the CAPN14 gene locus, demonstrate a molecular and cellular pathway that contributes to T helper type 2 responses in mucosal epithelium.
Figures






References
-
- Ono Y, Sorimachi H. Calpains: an elaborate proteolytic system. Biochim Biophys Acta. 2012;1824(1):224–236. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

