Genome-Wide Analysis Identifies Germ-Line Risk Factors Associated with Canine Mammary Tumours
- PMID: 27158822
- PMCID: PMC4861258
- DOI: 10.1371/journal.pgen.1006029
Genome-Wide Analysis Identifies Germ-Line Risk Factors Associated with Canine Mammary Tumours
Abstract
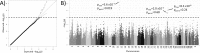
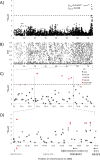
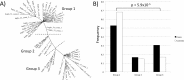
Canine mammary tumours (CMT) are the most common neoplasia in unspayed female dogs. CMTs are suitable naturally occurring models for human breast cancer and share many characteristics, indicating that the genetic causes could also be shared. We have performed a genome-wide association study (GWAS) in English Springer Spaniel dogs and identified a genome-wide significant locus on chromosome 11 (praw = 5.6x10-7, pperm = 0.019). The most associated haplotype spans a 446 kb region overlapping the CDK5RAP2 gene. The CDK5RAP2 protein has a function in cell cycle regulation and could potentially have an impact on response to chemotherapy treatment. Two additional loci, both on chromosome 27, were nominally associated (praw = 1.97x10-5 and praw = 8.30x10-6). The three loci explain 28.1±10.0% of the phenotypic variation seen in the cohort, whereas the top ten associated regions account for 38.2±10.8% of the risk. Furthermore, the ten GWAS loci and regions with reduced genetic variability are significantly enriched for snoRNAs and tumour-associated antigen genes, suggesting a role for these genes in CMT development. We have identified several candidate genes associated with canine mammary tumours, including CDK5RAP2. Our findings enable further comparative studies to investigate the genes and pathways in human breast cancer patients.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Torre LA, Siegel RL, Ward EM, Jemal A (2016) Global Cancer Incidence and Mortality Rates and Trends-An Update. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 25: 16–27. - PubMed
-
- Pharoah PD, Dunning AM, Ponder BA, Easton DF (2004) Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer 4: 850–860. - PubMed
-
- Claus EB, Schildkraut JM, Thompson WD, Risch NJ (1996) The genetic attributable risk of breast and ovarian cancer. Cancer 77: 2318–2324. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

