The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model
- PMID: 27160191
- PMCID: PMC4860760
- DOI: 10.1186/s12915-016-0258-1
The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model
Abstract
Background: Host-microbe associations underlie many key processes of host development, immunity, and life history. Yet, none of the current research on the central model species Caenorhabditis elegans considers the worm's natural microbiome. Instead, almost all laboratories exclusively use the canonical strain N2 and derived mutants, maintained through routine bleach sterilization in monoxenic cultures with an E. coli strain as food. Here, we characterize for the first time the native microbiome of C. elegans and assess its influence on nematode life history characteristics.
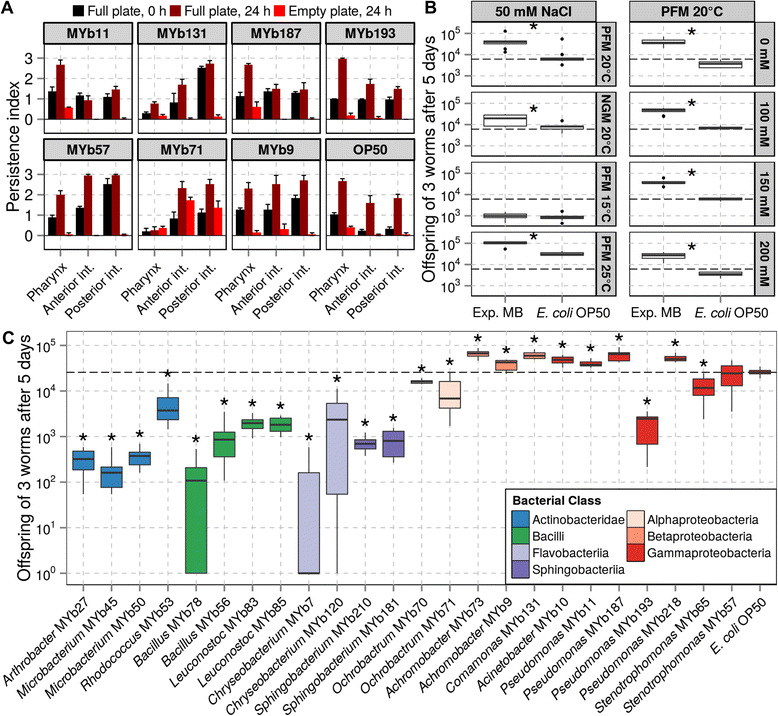
Results: Nematodes sampled directly from their native habitats carry a species-rich bacterial community, dominated by Proteobacteria such as Enterobacteriaceae and members of the genera Pseudomonas, Stenotrophomonas, Ochrobactrum, and Sphingomonas. The C. elegans microbiome is distinct from that of the worm's natural environment and the congeneric species C. remanei. Exposure to a derived experimental microbiome revealed that bacterial composition is influenced by host developmental stage and genotype. These experiments also showed that the microbes enhance host fitness under standard and also stressful conditions (e.g., high temperature and either low or high osmolarity). Taking advantage of the nematode's transparency, we further demonstrate that several Proteobacteria are able to enter the C. elegans gut and that an Ochrobactrum isolate even seems to be able to persist in the intestines under stressful conditions. Moreover, three Pseudomonas isolates produce an anti-fungal effect in vitro which we show can contribute to the worm's defense against fungal pathogens in vivo.
Conclusion: This first systematic analysis of the nematode's native microbiome reveals a species-rich bacterial community to be associated with C. elegans, which is likely of central importance for our understanding of the worm's biology. The information acquired and the microbial isolates now available for experimental work establishes C. elegans as a tractable model for the in-depth dissection of host-microbiome interactions.
Keywords: Antifungal defense; Caenorhabditis elegans; Holobiont; Metaorganism; Microbiome; Ochrobactrum; Pseudomonas; Sphingomonas; Stenotrophomonas.
Figures



Comment in
-
Caenorhabditis microbiota: worm guts get populated.BMC Biol. 2016 May 9;14:37. doi: 10.1186/s12915-016-0260-7. BMC Biol. 2016. PMID: 27160265 Free PMC article.
References
-
- Lederberg J, Mccray A. The Scientist: ‘Ome Sweet’ Omics–A Genealogical Treasury of Words. The Scientist. 2001;17:13313.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

