Rapid separation of bacteria from blood-review and outlook
- PMID: 27160415
- PMCID: PMC5297886
- DOI: 10.1002/btpr.2299
Rapid separation of bacteria from blood-review and outlook
Abstract
The high morbidity and mortality rate of bloodstream infections involving antibiotic-resistant bacteria necessitate a rapid identification of the infectious organism and its resistance profile. Traditional methods based on culturing the blood typically require at least 24 h, and genetic amplification by PCR in the presence of blood components has been problematic. The rapid separation of bacteria from blood would facilitate their genetic identification by PCR or other methods so that the proper antibiotic regimen can quickly be selected for the septic patient. Microfluidic systems that separate bacteria from whole blood have been developed, but these are designed to process only microliter quantities of whole blood or only highly diluted blood. However, symptoms of clinical blood infections can be manifest with bacterial burdens perhaps as low as 10 CFU/mL, and thus milliliter quantities of blood must be processed to collect enough bacteria for reliable genetic analysis. This review considers the advantages and shortcomings of various methods to separate bacteria from blood, with emphasis on techniques that can be done in less than 10 min on milliliter-quantities of whole blood. These techniques include filtration, screening, centrifugation, sedimentation, hydrodynamic focusing, chemical capture on surfaces or beads, field-flow fractionation, and dielectrophoresis. Techniques with the most promise include screening, sedimentation, and magnetic bead capture, as they allow large quantities of blood to be processed quickly. Some microfluidic techniques can be scaled up. © 2016 American Institute of Chemical Engineers Biotechnol. Prog., 32:823-839, 2016.
Keywords: bacterial bloodstream infection; centrifugation; chemical binding; filtration; hydrodynamic focusing; rapid identification; sedimentation.
© 2016 American Institute of Chemical Engineers.
Figures


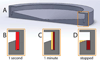
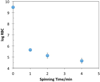
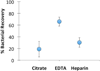

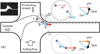

References
-
- CDC. [Accessed May 18, 2016];Antibiotic Resistance Threats in the United States, 2013. 2013 http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013....
-
- Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis. 2003;36:1418–1423. - PubMed
-
- Malik S, Ravishekhar K. Significance of coagulase negative Staphylococcus species in blood culture. J Clin Diagn Res. 2012;6:632–635.
-
- Howell L. Global Risks 2013: An Initiative of the Risk Response Network. New York, NY: World Economics Forum; 2013.
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

