Antimicrobial combinations: Bliss independence and Loewe additivity derived from mechanistic multi-hit models
- PMID: 27160596
- PMCID: PMC4874391
- DOI: 10.1098/rstb.2015.0294
Antimicrobial combinations: Bliss independence and Loewe additivity derived from mechanistic multi-hit models
Abstract
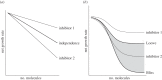
Antimicrobial peptides (AMPs) and antibiotics reduce the net growth rate of bacterial populations they target. It is relevant to understand if effects of multiple antimicrobials are synergistic or antagonistic, in particular for AMP responses, because naturally occurring responses involve multiple AMPs. There are several competing proposals describing how multiple types of antimicrobials add up when applied in combination, such as Loewe additivity or Bliss independence. These additivity terms are defined ad hoc from abstract principles explaining the supposed interaction between the antimicrobials. Here, we link these ad hoc combination terms to a mathematical model that represents the dynamics of antimicrobial molecules hitting targets on bacterial cells. In this multi-hit model, bacteria are killed when a certain number of targets are hit by antimicrobials. Using this bottom-up approach reveals that Bliss independence should be the model of choice if no interaction between antimicrobial molecules is expected. Loewe additivity, on the other hand, describes scenarios in which antimicrobials affect the same components of the cell, i.e. are not acting independently. While our approach idealizes the dynamics of antimicrobials, it provides a conceptual underpinning of the additivity terms. The choice of the additivity term is essential to determine synergy or antagonism of antimicrobials.This article is part of the themed issue 'Evolutionary ecology of arthropod antimicrobial peptides'.
Keywords: antagonism; antibiotics; antimicrobial peptide responses; mathematical modelling; pharmacodynamic function; synergy.
© 2016 The Author(s).
Figures
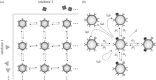
 and to class N1,2 with the rate
and to class N1,2 with the rate  Cells from class N0,1 transit to N1,1 with the rate
Cells from class N0,1 transit to N1,1 with the rate  and from class N1,0 with the rate
and from class N1,0 with the rate  . Inhibitors detach from the cells in class N1,1 at the rates
. Inhibitors detach from the cells in class N1,1 at the rates  and
and  respectively. The class N1,1 gains cells through detachment of inhibitors from cells of higher classes with
respectively. The class N1,1 gains cells through detachment of inhibitors from cells of higher classes with  in the case of detachment of inhibitor 1 and
in the case of detachment of inhibitor 1 and  in the case of detachment of inhibitor 2. The cells replicate at the rate b1,1N1,1; of all cells replicated, one-fourth stay in the class N1,1 whereas each one-fourth of the cells are categorized to the classes N0,1, N1,0 and N0,0, respectively. Lastly, cells can die at the rate d1,1
N1,1. Because we assumed the parameter to be the same independent of the class they are in, we use the following simplified expressions for the parameters: α1 instead of α1,1,1 and α1,2,1, α2 instead of α2,1,1 and α2,1,2, μ1 instead of μ1,1,1 and μ1,2,1, μ2 instead of μ2,1,1 and μ2,1,2, b instead of b1,1, and d instead of d1,1.
in the case of detachment of inhibitor 2. The cells replicate at the rate b1,1N1,1; of all cells replicated, one-fourth stay in the class N1,1 whereas each one-fourth of the cells are categorized to the classes N0,1, N1,0 and N0,0, respectively. Lastly, cells can die at the rate d1,1
N1,1. Because we assumed the parameter to be the same independent of the class they are in, we use the following simplified expressions for the parameters: α1 instead of α1,1,1 and α1,2,1, α2 instead of α2,1,1 and α2,1,2, μ1 instead of μ1,1,1 and μ1,2,1, μ2 instead of μ2,1,1 and μ2,1,2, b instead of b1,1, and d instead of d1,1.

Similar articles
-
Bacterial strategies of resistance to antimicrobial peptides.Philos Trans R Soc Lond B Biol Sci. 2016 May 26;371(1695):20150292. doi: 10.1098/rstb.2015.0292. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27160595 Free PMC article. Review.
-
Perspectives on the evolutionary ecology of arthropod antimicrobial peptides.Philos Trans R Soc Lond B Biol Sci. 2016 May 26;371(1695):20150297. doi: 10.1098/rstb.2015.0297. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27160599 Free PMC article. Review.
-
Diversity, evolution and medical applications of insect antimicrobial peptides.Philos Trans R Soc Lond B Biol Sci. 2016 May 26;371(1695):20150290. doi: 10.1098/rstb.2015.0290. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27160593 Free PMC article. Review.
-
Application of Antimicrobial Peptides of the Innate Immune System in Combination With Conventional Antibiotics-A Novel Way to Combat Antibiotic Resistance?Front Cell Infect Microbiol. 2019 Apr 30;9:128. doi: 10.3389/fcimb.2019.00128. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31114762 Free PMC article.
-
Friend, foe or food? Recognition and the role of antimicrobial peptides in gut immunity and Drosophila-microbe interactions.Philos Trans R Soc Lond B Biol Sci. 2016 May 26;371(1695):20150295. doi: 10.1098/rstb.2015.0295. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27160597 Free PMC article. Review.
Cited by
-
Additivity of inhibitory effects in multidrug combinations.Nat Microbiol. 2018 Dec;3(12):1339-1345. doi: 10.1038/s41564-018-0252-1. Epub 2018 Oct 15. Nat Microbiol. 2018. PMID: 30323252 Free PMC article.
-
Novel Synergies and Isolate Specificities in the Drug Interaction Landscape of Mycobacterium abscessus.Antimicrob Agents Chemother. 2023 Jul 18;67(7):e0009023. doi: 10.1128/aac.00090-23. Epub 2023 Jun 6. Antimicrob Agents Chemother. 2023. PMID: 37278639 Free PMC article.
-
Interactions of plumbagin with five common antibiotics against Staphylococcus aureus in vitro.PLoS One. 2024 Jan 26;19(1):e0297493. doi: 10.1371/journal.pone.0297493. eCollection 2024. PLoS One. 2024. PMID: 38277418 Free PMC article.
-
Pyoverdine-antibiotic combination treatment: its efficacy and effects on resistance evolution in Escherichia coli.Microlife. 2024 Oct 15;5:uqae021. doi: 10.1093/femsml/uqae021. eCollection 2024. Microlife. 2024. PMID: 39502382 Free PMC article.
-
Bliss' and Loewe's additive and synergistic effects in Plasmodium falciparum growth inhibition by AMA1-RON2L, RH5, RIPR and CyRPA antibody combinations.Sci Rep. 2020 Jul 16;10(1):11802. doi: 10.1038/s41598-020-67877-8. Sci Rep. 2020. PMID: 32678144 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

