A comprehensive survey of the mutagenic impact of common cancer cytotoxics
- PMID: 27161042
- PMCID: PMC4862131
- DOI: 10.1186/s13059-016-0963-7
A comprehensive survey of the mutagenic impact of common cancer cytotoxics
Abstract
Background: Genomic mutations caused by cytotoxic agents used in cancer chemotherapy may cause secondary malignancies as well as contribute to the evolution of treatment-resistant tumour cells. The stable diploid genome of the chicken DT40 lymphoblast cell line, an established DNA repair model system, is well suited to accurately assay genomic mutations.
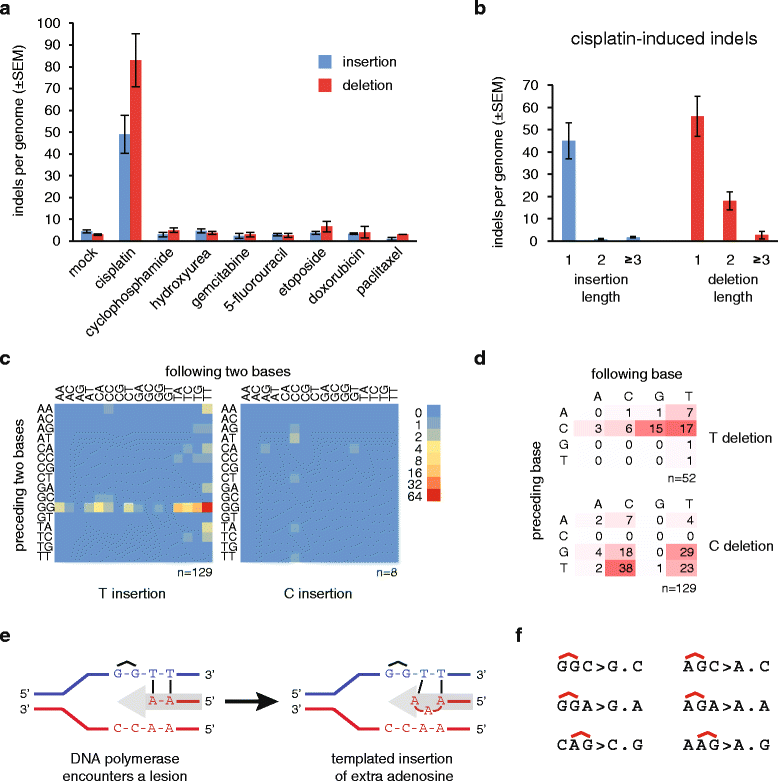
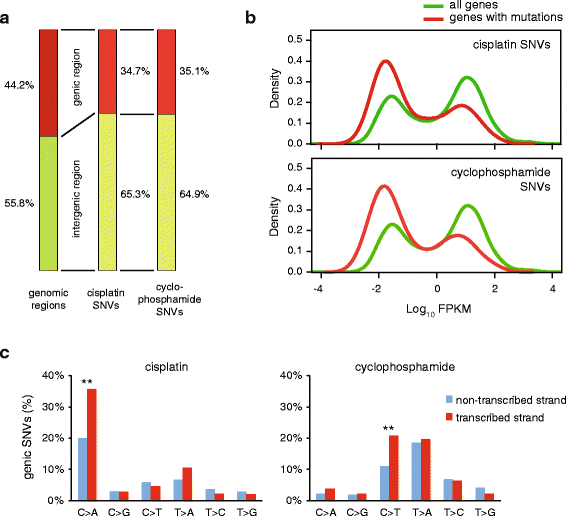
Results: We use whole genome sequencing of multiple DT40 clones to determine the mutagenic effect of eight common cytotoxics used for the treatment of millions of patients worldwide. We determine the spontaneous mutagenesis rate at 2.3 × 10(-10) per base per cell division and find that cisplatin, cyclophosphamide and etoposide induce extra base substitutions with distinct spectra. After four cycles of exposure, cisplatin induces 0.8 mutations per Mb, equivalent to the median mutational burden in common leukaemias. Cisplatin-induced mutations, including short insertions and deletions, are mainly located at sites of putative intrastrand crosslinks. We find two of the newly defined cisplatin-specific mutation types as causes of the reversion of BRCA2 mutations in emerging cisplatin-resistant tumours or cell clones. Gemcitabine, 5-fluorouracil, hydroxyurea, doxorubicin and paclitaxel have no measurable mutagenic effect. The cisplatin-induced mutation spectrum shows good correlation with cancer mutation signatures attributed to smoking and other sources of guanine-directed base damage.
Conclusion: This study provides support for the use of cell line mutagenesis assays to validate or predict the mutagenic effect of environmental and iatrogenic exposures. Our results suggest genetic reversion due to cisplatin-induced mutations as a distinct mechanism for developing resistance.
Keywords: BRCA2; Cancer chemotherapy; Chemotherapy resistance; Cisplatin; Cyclophosphamide; Cytotoxics; DT40; Etoposide; Mutagenesis; Spontaneous mutagenesis; Whole genome sequencing.
Figures




References
-
- OECD Guidelines for the Testing of Chemicals, Section 4:Health Effects. OECD Publications. 2015. http://www.oecd-ilibrary.org/environment/oecd-guidelines-for-the-testing.... Accessed 12 Nov 2015
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

