Novel exon 1 protein-coding regions N-terminally extend human KCNE3 and KCNE4
- PMID: 27162025
- PMCID: PMC6137956
- DOI: 10.1096/fj.201600467R
Novel exon 1 protein-coding regions N-terminally extend human KCNE3 and KCNE4
Abstract
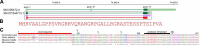
The 5 human (h)KCNE β subunits each regulate various cation channels and are linked to inherited cardiac arrhythmias. Reported here are previously undiscovered protein-coding regions in exon 1 of hKCNE3 and hKCNE4 that extend their encoded extracellular domains by 44 and 51 residues, which yields full-length proteins of 147 and 221 residues, respectively. Full-length hKCNE3 and hKCNE4 transcript and protein are expressed in multiple human tissues; for hKCNE4, only the longer protein isoform is detectable. Two-electrode voltage-clamp electrophysiology revealed that, when coexpressed in Xenopus laevis oocytes with various potassium channels, the newly discovered segment preserved conversion of KCNQ1 by hKCNE3 to a constitutively open channel, but prevented its inhibition of Kv4.2 and KCNQ4. hKCNE4 slowing of Kv4.2 inactivation and positive-shifted steady-state inactivation were also preserved in the longer form. In contrast, full-length hKCNE4 inhibition of KCNQ1 was limited to 40% at +40 mV vs. 80% inhibition by the shorter form, and augmentation of KCNQ4 activity by hKCNE4 was entirely abolished by the additional segment. Among the genome databases analyzed, the longer KCNE3 is confined to primates; full-length KCNE4 is widespread in vertebrates but is notably absent from Mus musculus Findings highlight unexpected KCNE gene diversity, raise the possibility of dynamic regulation of KCNE partner modulation via splice variation, and suggest that the longer hKCNE3 and hKCNE4 proteins should be adopted in future mechanistic and genetic screening studies.-Abbott, G. W. Novel exon 1 protein-coding regions N-terminally extend human KCNE3 and KCNE4.
Keywords: KCNQ1; KCNQ4; Kv4.2; arrhythmia; potassium channel.
© FASEB.
Conflict of interest statement
The author is grateful for helpful discussions with Dr. Alex Mercer, Daniel Neverisky (University of California, Irvine) and Dr. Nancy Carrasco (Yale University, New Haven, CT, USA). The author thanks Dr. David Christini and Lala Tanmoy Das (Weill Cornell Medical College, New York, NY, USA) for invaluable tissue samples. This work was supported by the U.S. National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases Grant DK41544, and National Institute of General Medical Sciences Grant GM115189).
Figures









References
-
- Isacoff E. Y., Jan Y. N., Jan L. Y. (1990) Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature , 530–534 - PubMed
-
- Abbott G. W., Goldstein S. A. (1998) A superfamily of small potassium channel subunits: form and function of the MinK-related peptides (MiRPs). Q. Rev. Biophys. , 357–398 - PubMed
-
- McCrossan Z. A., Abbott G. W. (2004) The MinK-related peptides. Neuropharmacology , 787–821 - PubMed
-
- Abbott G. W., Butler M. H., Bendahhou S., Dalakas M. C., Ptacek L. J., Goldstein S. A. (2001) MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell , 217–231 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

