Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort
- PMID: 27162351
- PMCID: PMC4890596
- DOI: 10.1073/pnas.1603712113
Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort
Abstract
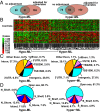
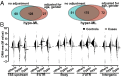
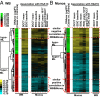
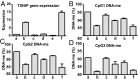
We examined whether persistence of epigenetic DNA methylation (DNA-me) alterations at specific loci over two different time points in people with diabetes are associated with metabolic memory, the prolonged beneficial effects of intensive vs. conventional therapy during the Diabetes Control and Complications Trial (DCCT) on the progression of microvascular outcomes in the long-term follow-up Epidemiology of Diabetes Interventions and Complications (EDIC) Study. We compared DNA-me profiles in genomic DNA of whole blood (WB) isolated at EDIC Study baseline from 32 cases (DCCT conventional therapy group subjects showing retinopathy or albuminuria progression by EDIC Study year 10) vs. 31 controls (DCCT intensive therapy group subjects without complication progression by EDIC year 10). DNA-me was also profiled in blood monocytes (Monos) of the same patients obtained during EDIC Study years 16-17. In WB, 153 loci depicted hypomethylation, and 225 depicted hypermethylation, whereas in Monos, 155 hypomethylated loci and 247 hypermethylated loci were found (fold change ≥1.3; P < 0.005; cases vs. controls). Twelve annotated differentially methylated loci were common in both WB and Monos, including thioredoxin-interacting protein (TXNIP), known to be associated with hyperglycemia and related complications. A set of differentially methylated loci depicted similar trends of associations with prior HbA1c in both WB and Monos. In vitro, high glucose induced similar persistent hypomethylation at TXNIP in cultured THP1 Monos. These results show that DNA-me differences during the DCCT persist at certain loci associated with glycemia for several years during the EDIC Study and support an epigenetic explanation for metabolic memory.
Keywords: DNA methylation; TXNIP; diabetic complications; epigenetics; metabolic memory.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ihnat MA, Thorpe JE, Ceriello A. Hypothesis: The ‘metabolic memory’, the new challenge of diabetes. Diabet Med. 2007;24(6):582–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

