Candidate gene networks and blood biomarkers of methamphetamine-associated psychosis: an integrative RNA-sequencing report
- PMID: 27163203
- PMCID: PMC5070070
- DOI: 10.1038/tp.2016.67
Candidate gene networks and blood biomarkers of methamphetamine-associated psychosis: an integrative RNA-sequencing report
Abstract
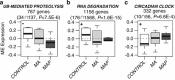
The clinical presentation, course and treatment of methamphetamine (METH)-associated psychosis (MAP) are similar to that observed in schizophrenia (SCZ) and subsequently MAP has been hypothesized as a pharmacological and environmental model of SCZ. However, several challenges currently exist in diagnosing MAP accurately at the molecular and neurocognitive level before the MAP model can contribute to the discovery of SCZ biomarkers. We directly assessed subcortical brain structural volumes and clinical parameters of MAP within the framework of an integrative genome-wide RNA-Seq blood transcriptome analysis of subjects diagnosed with MAP (N=10), METH dependency without psychosis (MA; N=10) and healthy controls (N=10). First, we identified discrete groups of co-expressed genes (that is, modules) and tested them for functional annotation and phenotypic relationships to brain structure volumes, life events and psychometric measurements. We discovered one MAP-associated module involved in ubiquitin-mediated proteolysis downregulation, enriched with 61 genes previously found implicated in psychosis and SCZ across independent blood and post-mortem brain studies using convergent functional genomic (CFG) evidence. This module demonstrated significant relationships with brain structure volumes including the anterior corpus callosum (CC) and the nucleus accumbens. Furthermore, a second MAP and psychoticism-associated module involved in circadian clock upregulation was also enriched with 39 CFG genes, further associated with the CC. Subsequently, a machine-learning analysis of differentially expressed genes identified single blood-based biomarkers able to differentiate controls from methamphetamine dependents with 87% accuracy and MAP from MA subjects with 95% accuracy. CFG evidence validated a significant proportion of these putative MAP biomarkers in independent studies including CLN3, FBP1, TBC1D2 and ZNF821 (RNA degradation), ELK3 and SINA3 (circadian clock) and PIGF and UHMK1 (ubiquitin-mediated proteolysis). Finally, focusing analysis on brain structure volumes revealed significantly lower bilateral hippocampal volumes in MAP subjects. Overall, these results suggest similar molecular and neurocognitive mechanisms underlying the pathophysiology of psychosis and SCZ regardless of substance abuse and provide preliminary evidence supporting the MAP paradigm as an exemplar for SCZ biomarker discovery.
Figures


References
-
- Yang MH, Jung MS, Lee MJ, Yoo KH, Yook YJ, Park EY et al. Gene expression profiling of the rewarding effect caused by methamphetamine in the mesolimbic dopamine system. Mol Cells 2008; 26: 121–130. - PubMed
-
- United Nations Office on Drugs and Crime. World Drug Report 2004UN Office on Drugs and Crime: Vienna, Austria.
-
- Kapp C. Crystal meth boom adds to South Africa's health challenges. Lancet 2008; 371: 193–194. - PubMed
-
- Nutt DJ, King LA, Phillips LD. Drug harms in the UK: a multicriteria decision analysis. Lancet 2010; 376: 1558–1565. - PubMed
-
- Srisurapanont M, Ali R, Marsden J, Sunga A, Wada K, Monteiro M. Psychotic symptoms in methamphetamine psychotic in-patients. Int J Neuropsychopharmacol 2003; 6: 347–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

