Differential expression of transcriptional regulatory units in the prefrontal cortex of patients with bipolar disorder: potential role of early growth response gene 3
- PMID: 27163206
- PMCID: PMC5070056
- DOI: 10.1038/tp.2016.78
Differential expression of transcriptional regulatory units in the prefrontal cortex of patients with bipolar disorder: potential role of early growth response gene 3
Abstract
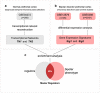
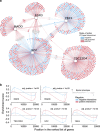
Bipolar disorder (BD) is a severe mental illness with a strong genetic component. Despite its high degree of heritability, current genetic studies have failed to reveal individual loci of large effect size. In lieu of focusing on individual genes, we investigated regulatory units (regulons) in BD to identify candidate transcription factors (TFs) that regulate large groups of differentially expressed genes. Network-based approaches should elucidate the molecular pathways governing the pathophysiology of BD and reveal targets for potential therapeutic intervention. The data from a large-scale microarray study was used to reconstruct the transcriptional associations in the human prefrontal cortex, and results from two independent microarray data sets to obtain BD gene signatures. The regulatory network was derived by mapping the significant interactions between known TFs and all potential targets. Five regulons were identified in both transcriptional network models: early growth response 3 (EGR3), TSC22 domain family, member 4 (TSC22D4), interleukin enhancer-binding factor 2 (ILF2), Y-box binding protein 1 (YBX1) and MAP-kinase-activating death domain (MADD). With a high stringency threshold, the consensus across tests was achieved only for the EGR3 regulon. We identified EGR3 in the prefrontal cortex as a potential key target, robustly repressed in both BD signatures. Considering that EGR3 translates environmental stimuli into long-term changes in the brain, disruption in biological pathways involving EGR3 may induce an impaired response to stress and influence on risk for psychiatric disorders, particularly BD.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

