Computational cell analysis for label-free detection of cell properties in a microfluidic laminar flow
- PMID: 27163941
- PMCID: PMC4914432
- DOI: 10.1039/c6an00295a
Computational cell analysis for label-free detection of cell properties in a microfluidic laminar flow
Abstract
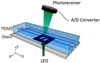
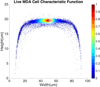
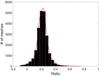
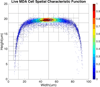
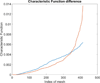
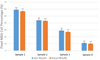
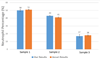
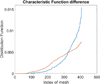
Although a flow cytometer, being one of the most popular research and clinical tools for biomedicine, can analyze cells based on the cell size, internal structures such as granularity, and molecular markers, it provides little information about the physical properties of cells such as cell stiffness and physical interactions between the cell membrane and fluid. In this paper, we propose a computational cell analysis technique using cells' different equilibrium positions in a laminar flow. This method utilizes a spatial coding technique to acquire the spatial position of the cell in a microfluidic channel and then uses mathematical algorithms to calculate the ratio of cell mixtures. Most uniquely, the invented computational cell analysis technique can unequivocally detect the subpopulation of each cell type without labeling even when the cell type shows a substantial overlap in the distribution plot with other cell types, a scenario limiting the use of conventional flow cytometers and machine learning techniques. To prove this concept, we have applied the computation method to distinguish live and fixed cancer cells without labeling, count neutrophils from human blood, and distinguish drug treated cells from untreated cells. Our work paves the way for using computation algorithms and fluidic dynamic properties for cell classification, a label-free method that can potentially classify over 200 types of human cells. Being a highly cost-effective cell analysis method complementary to flow cytometers, our method can offer orthogonal tests in companion with flow cytometers to provide crucial information for biomedical samples.
Figures





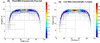

References
-
- Pang Lin, et al. Optofluidic devices and applications in photonics, sensing and imaging. Lab on a Chip. 2012;12.19:3543–3551. - PubMed
-
- Wu Jigang, Zheng Guoan, Lee Lap Man. Optical imaging techniques in microfluidics and their applications. Lab on a Chip. 2012;12.19:3566–3575. - PubMed
-
- Monat C, Domachuk P, Eggleton BJ. Integrated optofluidics: A new river of light. Nature photonics. 2007;1.2:106–114.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

