Cell-free translation and purification of Arabidopsis thaliana regulator of G signaling 1 protein
- PMID: 27164033
- PMCID: PMC5594927
- DOI: 10.1016/j.pep.2016.04.016
Cell-free translation and purification of Arabidopsis thaliana regulator of G signaling 1 protein
Abstract
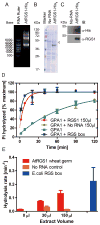
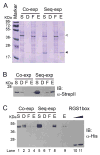
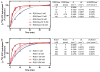
Arabidopsis thaliana Regulator of G protein Signalling 1 (AtRGS1) is a protein with a predicted N-terminal 7-transmembrane (7TM) domain and a C-terminal cytosolic RGS1 box domain. The RGS1 box domain exerts GTPase activation (GAP) activity on Gα (AtGPA1), a component of heterotrimeric G protein signaling in plants. AtRGS1 may perceive an exogenous agonist to regulate the steady-state levels of the active form of AtGPA1. It is uncertain if the full-length AtRGS1 protein exerts any atypical effects on Gα, nor has it been established exactly how AtRGS1 contributes to perception of an extracellular signal and transmits this response to a G-protein dependent signaling cascade. Further studies on full-length AtRGS1 have been inhibited due to the extreme low abundance of the endogenous AtRGS1 protein in plants and lack of a suitable heterologous system to express AtRGS1. Here, we describe methods to produce full-length AtRGS1 by cell-free synthesis into unilamellar liposomes and nanodiscs. The cell-free synthesized AtRGS1 exhibits GTPase activating activity on Gα and can be purified to a level suitable for biochemical analyses.
Keywords: 7-Transmembrane protein; Arabidopsis regulator of G signaling protein 1 (AtRGS1); In vitro translation; Membrane scaffold protein 1D1; Nanodiscs.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Purification and in vitro functional analysis of the Arabidopsis thaliana regulator of G-protein signaling-1.Methods Enzymol. 2004;389:320-38. doi: 10.1016/S0076-6879(04)89019-0. Methods Enzymol. 2004. PMID: 15313574 Review.
-
AtRGS1 function in Arabidopsis thaliana.Methods Enzymol. 2004;389:338-50. doi: 10.1016/S0076-6879(04)89020-7. Methods Enzymol. 2004. PMID: 15313575 Review.
-
Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana.J Biol Chem. 2011 Apr 15;286(15):13143-50. doi: 10.1074/jbc.M110.190355. Epub 2011 Feb 16. J Biol Chem. 2011. PMID: 21325279 Free PMC article.
-
The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination.Plant Physiol. 2006 Jan;140(1):302-10. doi: 10.1104/pp.105.069872. Epub 2005 Dec 16. Plant Physiol. 2006. PMID: 16361523 Free PMC article.
-
D-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1.FEBS Lett. 2008 Oct 29;582(25-26):3577-84. doi: 10.1016/j.febslet.2008.08.038. Epub 2008 Sep 24. FEBS Lett. 2008. PMID: 18817773 Free PMC article.
Cited by
-
Direct Modulation of Heterotrimeric G Protein-coupled Signaling by a Receptor Kinase Complex.J Biol Chem. 2016 Jul 1;291(27):13918-13925. doi: 10.1074/jbc.C116.736702. Epub 2016 May 27. J Biol Chem. 2016. PMID: 27235398 Free PMC article.
-
A cell-free method for expressing and reconstituting membrane proteins enables functional characterization of the plant receptor-like protein kinase FERONIA.J Biol Chem. 2017 Apr 7;292(14):5932-5942. doi: 10.1074/jbc.M116.761981. Epub 2017 Feb 24. J Biol Chem. 2017. PMID: 28235802 Free PMC article.
-
Easy Synthesis of Complex Biomolecular Assemblies: Wheat Germ Cell-Free Protein Expression in Structural Biology.Front Mol Biosci. 2021 Mar 25;8:639587. doi: 10.3389/fmolb.2021.639587. eCollection 2021. Front Mol Biosci. 2021. PMID: 33842544 Free PMC article. Review.
-
Nanodiscs in Membrane Biochemistry and Biophysics.Chem Rev. 2017 Mar 22;117(6):4669-4713. doi: 10.1021/acs.chemrev.6b00690. Epub 2017 Feb 8. Chem Rev. 2017. PMID: 28177242 Free PMC article. Review.
-
Half a Century of Progress: The Evolution of Wheat Germ-Based In Vitro Translation into a Versatile Protein Production Method.Int J Mol Sci. 2025 Apr 10;26(8):3577. doi: 10.3390/ijms26083577. Int J Mol Sci. 2025. PMID: 40332070 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

