Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection
- PMID: 27164078
- PMCID: PMC4881435
- DOI: 10.3390/ijms17050585
Specific Magnetic Isolation of E6 HPV16 Modified Magnetizable Particles Coupled with PCR and Electrochemical Detection
Abstract
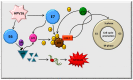
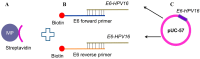
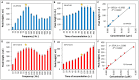
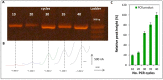
The majority of carcinomas that were developed due to the infection with human papillomavirus (HPV) are caused by high-risk HPV types, HPV16 and HPV18. These HPV types contain the E6 and E7 oncogenes, so the fast detection of these oncogenes is an important point to avoid the development of cancer. Many different HPV tests are available to detect the presence of HPV in biological samples. The aim of this study was to design a fast and low cost method for HPV identification employing magnetic isolation, polymerase chain reaction (PCR) and electrochemical detection. These assays were developed to detect the interactions between E6-HPV16 oncogene and magnetizable particles (MPs) using commercial Dynabeads M-280 Streptavidin particles and laboratory-synthesized "homemade" particles called MANs (MAN-37, MAN-127 and MAN-164). The yields of PCR amplification of E6-HPV16 oncogene bound on the particles and after the elution from the particles were compared. A highest yield of E6-HPV16 DNA isolation was obtained with both MPs particles commercial M-280 Streptavidin and MAN-37 due to reducing of the interferents compared with the standard PCR method. A biosensor employing the isolation of E6-HPV16 oncogene with MPs particles followed by its electrochemical detection can be a very effective technique for HPV identification, providing simple, sensitive and cost-effective analysis.
Keywords: PCR; electrochemistry; human papillomavirus; magnetic isolation; magnetizable particles; nucleic acid detection.
Figures





Similar articles
-
Human papillomavirus infections and upper aero-digestive tract cancers: the ARCAGE study.J Natl Cancer Inst. 2013 Apr 17;105(8):536-45. doi: 10.1093/jnci/djt053. Epub 2013 Mar 16. J Natl Cancer Inst. 2013. PMID: 23503618
-
Α 2-stage, nested-like nucleic acid amplification method (IsoPCR) for the highly sensitive detection of HPV16 and HPV18 DNA.Mol Cell Probes. 2019 Jun;45:1-7. doi: 10.1016/j.mcp.2019.03.003. Epub 2019 Mar 19. Mol Cell Probes. 2019. PMID: 30902662
-
Distinctive distribution of HPV16 E6 D25E and E7 N29S intratypic Asian variants in Korean commercial sex workers.J Med Virol. 2007 Apr;79(4):426-30. doi: 10.1002/jmv.20826. J Med Virol. 2007. PMID: 17311337
-
[Gene characterization of E6 and E7 gene of human papillomavirus of 15 cervical cancer in Beijing].Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2009 Apr;23(2):88-90. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2009. PMID: 20104743 Chinese.
-
HPV prevalence, E6 sequence variation and physical state of HPV16 isolates from patients with cervical cancer in Sichuan, China.Gynecol Oncol. 2007 Jan;104(1):77-85. doi: 10.1016/j.ygyno.2006.07.016. Epub 2006 Sep 12. Gynecol Oncol. 2007. PMID: 16970982
Cited by
-
Electrochemical Duplex Detection of E2 and E6 Genes of Human Papillomavirus Type 16 and Determination of Physical Status in High-Risk Cervical Carcinoma.J Med Virol. 2025 Mar;97(3):e70299. doi: 10.1002/jmv.70299. J Med Virol. 2025. PMID: 40071579 Free PMC article.
-
Long intergenic non-coding LINC00657 regulates tumorigenesis of glioblastoma by acting as a molecular sponge of miR-190a-3p.Aging (Albany NY). 2019 Mar 5;11(5):1456-1470. doi: 10.18632/aging.101845. Aging (Albany NY). 2019. PMID: 30837348 Free PMC article.
-
Nanoparticles for Signaling in Biodiagnosis and Treatment of Infectious Diseases.Int J Mol Sci. 2018 May 31;19(6):1627. doi: 10.3390/ijms19061627. Int J Mol Sci. 2018. PMID: 29857492 Free PMC article. Review.
References
-
- Parfenov M., Pedamallu C.S., Gehlenborg N., Freeman S.S., Danilova L., Bristow C.A., Lee S., Hadjipanayis A.G., Ivanova E.V., Wilkerson M.D., et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. - DOI - PMC - PubMed
-
- Ruttkay-Nedecky B., Jimenez A.M.J., Nejdl L., Chudobova D., Gumulec J., Masarik M., Adam V., Kizek R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins. Int. J. Oncol. 2013;43:1754–1762. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

