Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors
- PMID: 27164086
- PMCID: PMC4881508
- DOI: 10.3390/ijms17050682
Functional and Activity Analysis of Cattle UCP3 Promoter with MRFs-Related Factors
Abstract
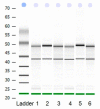
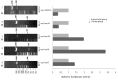
Uncoupling protein 3 (UCP3) is mainly expressed in muscle. It plays an important role in muscle, but less research on the regulation of cattle UCP3 has been performed. In order to elucidate whether cattle UCP3 can be regulated by muscle-related factors, deletion of cattle UCP3 promoter was amplified and cloned into pGL3-basic, pGL3-promoter and PEGFP-N3 vector, respectively, then transfected into C2C12 myoblasts cells and UCP3 promoter activity was measured using the dual-Luciferase reporter assay system. The results showed that there is some negative-regulatory element from -620 to -433 bp, and there is some positive-regulatory element between -433 and -385 bp. The fragment (1.08 kb) of UCP3 promoter was cotransfected with muscle-related transcription factor myogenic regulatory factors (MRFs) and myocyte-specific enhancer factor 2A (MEF2A). We found that UCP3 promoter could be upregulated by Myf5, Myf6 and MyoD and downregulated by MyoG and MEF2A.
Keywords: MRFs family; myocyte-specific enhancer factor 2A (MEF2A); promoter; transcriptional activity; uncoupling protein 3 (UCP3).
Figures









Similar articles
-
Differential regulation of the promoter activity of the mouse UCP2 and UCP3 genes by MyoD and myogenin.J Biochem Mol Biol. 2007 Nov 30;40(6):921-7. doi: 10.5483/bmbrep.2007.40.6.921. J Biochem Mol Biol. 2007. PMID: 18047787
-
Chicken ovalbumin upstream promoter transcription factor II regulates uncoupling protein 3 gene transcription in Phodopus sungorus.BMC Mol Biol. 2007 Jan 4;8:1. doi: 10.1186/1471-2199-8-1. BMC Mol Biol. 2007. PMID: 17204145 Free PMC article.
-
Functional analysis of pig myostatin gene promoter with some adipogenesis- and myogenesis-related factors.Mol Cell Biochem. 2012 Apr;363(1-2):291-9. doi: 10.1007/s11010-011-1181-y. Epub 2011 Dec 11. Mol Cell Biochem. 2012. PMID: 22160830
-
Functional relationship between MyoD and peroxisome proliferator-activated receptor-dependent regulatory pathways in the control of the human uncoupling protein-3 gene transcription.Mol Endocrinol. 2003 Oct;17(10):1944-58. doi: 10.1210/me.2002-0395. Epub 2003 Jul 3. Mol Endocrinol. 2003. PMID: 12843208
-
The Interplay of UCP3 and PCSK1 Variants in Severe Obesity.Curr Obes Rep. 2025 Apr 26;14(1):38. doi: 10.1007/s13679-025-00631-1. Curr Obes Rep. 2025. PMID: 40281302 Free PMC article. Review.
Cited by
-
An Identification of Functional Genetic Variants in B4GALNT2 Gene and Their Association with Growth Traits in Goats.Genes (Basel). 2024 Mar 3;15(3):330. doi: 10.3390/genes15030330. Genes (Basel). 2024. PMID: 38540389 Free PMC article.
-
O-GlcNAcylation modification of MyoD regulates skeletal muscle fiber differentiation by antagonizing the UPF1 pathway.J Biol Chem. 2025 Apr;301(4):108364. doi: 10.1016/j.jbc.2025.108364. Epub 2025 Feb 27. J Biol Chem. 2025. PMID: 40023397 Free PMC article.
-
Study on the transcriptional regulatory mechanism of the MyoD1 gene in Guanling bovine.RSC Adv. 2018 Apr 3;8(22):12409-12419. doi: 10.1039/c7ra11795g. eCollection 2018 Mar 26. RSC Adv. 2018. PMID: 35548782 Free PMC article.
References
-
- Solanes G., Pedraza N., Iglesias R., Giralt M., Villarroya F. Functional Relationship between MyoD and Peroxisome Proliferator-Activated Receptor Dependent Regulatory Pathways in the Control of the Human Uncoupling Protein-3 Gene Transcription. Mol. Endocrinol. 2003;17:1944–1958. doi: 10.1210/me.2002-0395. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

