Toxin-Antitoxin Systems of Staphylococcus aureus
- PMID: 27164142
- PMCID: PMC4885055
- DOI: 10.3390/toxins8050140
Toxin-Antitoxin Systems of Staphylococcus aureus
Abstract
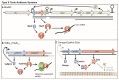
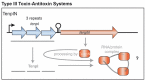
Toxin-antitoxin (TA) systems are small genetic elements found in the majority of prokaryotes. They encode toxin proteins that interfere with vital cellular functions and are counteracted by antitoxins. Dependent on the chemical nature of the antitoxins (protein or RNA) and how they control the activity of the toxin, TA systems are currently divided into six different types. Genes comprising the TA types I, II and III have been identified in Staphylococcus aureus. MazF, the toxin of the mazEF locus is a sequence-specific RNase that cleaves a number of transcripts, including those encoding pathogenicity factors. Two yefM-yoeB paralogs represent two independent, but auto-regulated TA systems that give rise to ribosome-dependent RNases. In addition, omega/epsilon/zeta constitutes a tripartite TA system that supposedly plays a role in the stabilization of resistance factors. The SprA1/SprA1AS and SprF1/SprG1 systems are post-transcriptionally regulated by RNA antitoxins and encode small membrane damaging proteins. TA systems controlled by interaction between toxin protein and antitoxin RNA have been identified in S. aureus in silico, but not yet experimentally proven. A closer inspection of possible links between TA systems and S. aureus pathophysiology will reveal, if these genetic loci may represent druggable targets. The modification of a staphylococcal TA toxin to a cyclopeptide antibiotic highlights the potential of TA systems as rather untapped sources of drug discovery.
Keywords: MazEF; Omega-Epsilon-Zeta; RNase; SprA1-SprA1AS; SprFG; Staphylococcus aureus; TenpIN; YefM-YoeB; plasmid addiction; toxin-antitoxin system.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

