The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events
- PMID: 27165002
- PMCID: PMC5027685
- DOI: 10.1038/ejhg.2016.40
The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events
Abstract
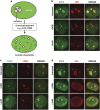
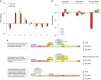
Ribosomal RNA synthesis occurs in the nucleolus and is a tightly regulated process that is targeted in some developmental diseases and hyperactivated in multiple cancers. Subcellular localization and immunoprecipitation coupled mass spectrometry demonstrated that a proportion of plant homeodomain (PHD) finger protein 6 (PHF6) protein is localized within the nucleolus and interacts with proteins involved in ribosomal processing. PHF6 sequence variants cause Börjeson-Forssman-Lehmann syndrome (BFLS, MIM#301900) and are also associated with a female-specific phenotype overlapping with Coffin-Siris syndrome (MIM#135900), T-cell acute lymphoblastic leukemia (MIM#613065), and acute myeloid leukemia (MIM#601626); however, very little is known about its cellular function, including its nucleolar role. HEK 293T cells were treated with RNase A, DNase I, actinomycin D, or 5,6-dichloro-β-D-ribofuranosylbenzimadole, followed by immunocytochemistry to determine PHF6 sub-nucleolar localization. We observed RNA-dependent localization of PHF6 to the sub-nucleolar fibrillar center (FC) and dense fibrillar component (DFC), at whose interface rRNA transcription occurs. Subsequent ChIP-qPCR analysis revealed strong enrichment of PHF6 across the entire rDNA-coding sequence but not along the intergenic spacer (IGS) region. When rRNA levels were quantified in a PHF6 gain-of-function model, we observed an overall decrease in rRNA transcription, accompanied by a modest increase in repressive promoter-associated RNA (pRNA) and a significant increase in the expression levels of the non-coding IGS36RNA and IGS39RNA transcripts. Collectively, our results demonstrate a role for PHF6 in carefully mediating the overall levels of ribosome biogenesis within a cell.
Figures


Similar articles
-
PHF6 functions as a tumor suppressor by recruiting methyltransferase SUV39H1 to nucleolar region and offers a novel therapeutic target for PHF6-muntant leukemia.Acta Pharm Sin B. 2022 Apr;12(4):1913-1927. doi: 10.1016/j.apsb.2021.10.025. Epub 2021 Oct 30. Acta Pharm Sin B. 2022. PMID: 35847518 Free PMC article.
-
PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex.J Proteome Res. 2012 Aug 3;11(8):4326-37. doi: 10.1021/pr3004369. Epub 2012 Jul 3. J Proteome Res. 2012. PMID: 22720776
-
PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis.J Biol Chem. 2013 Feb 1;288(5):3174-83. doi: 10.1074/jbc.M112.414839. Epub 2012 Dec 10. J Biol Chem. 2013. PMID: 23229552 Free PMC article.
-
Pathogenesis of Börjeson-Forssman-Lehmann syndrome: Insights from PHF6 function.Neurobiol Dis. 2016 Dec;96:227-235. doi: 10.1016/j.nbd.2016.09.011. Epub 2016 Sep 12. Neurobiol Dis. 2016. PMID: 27633282 Free PMC article. Review.
-
The Role of PHF6 in Hematopoiesis and Hematologic Malignancies.Stem Cell Rev Rep. 2023 Jan;19(1):67-75. doi: 10.1007/s12015-022-10447-4. Epub 2022 Aug 26. Stem Cell Rev Rep. 2023. PMID: 36008597 Review.
Cited by
-
Leukemia-mutated proteins PHF6 and PHIP form a chromatin complex that represses acute myeloid leukemia stemness.bioRxiv [Preprint]. 2024 Dec 18:2024.11.29.625909. doi: 10.1101/2024.11.29.625909. bioRxiv. 2024. Update in: Genes Dev. 2025 Jul 28. doi: 10.1101/gad.352602.125. PMID: 39677666 Free PMC article. Updated. Preprint.
-
Variability of Human rDNA.Cells. 2021 Jan 20;10(2):196. doi: 10.3390/cells10020196. Cells. 2021. PMID: 33498263 Free PMC article. Review.
-
Regulatory roles of nucleolus organizer region-derived long non-coding RNAs.Mamm Genome. 2022 Jun;33(2):402-411. doi: 10.1007/s00335-021-09906-z. Epub 2021 Aug 26. Mamm Genome. 2022. PMID: 34436664 Free PMC article. Review.
-
Cockayne syndrome group A and B proteins function in rRNA transcription through nucleolin regulation.Nucleic Acids Res. 2020 Mar 18;48(5):2473-2485. doi: 10.1093/nar/gkz1242. Nucleic Acids Res. 2020. PMID: 31970402 Free PMC article.
-
PHF6 cooperates with SWI/SNF complexes to facilitate transcriptional progression.Nat Commun. 2024 Aug 24;15(1):7303. doi: 10.1038/s41467-024-51566-5. Nat Commun. 2024. PMID: 39181868 Free PMC article.
References
-
- Lower KM, Turner G, Kerr BA et al: Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat Genet 2002; 32: 661–665. - PubMed
-
- Gecz J, Turner G, Nelson J, Partington M: The Borjeson-Forssman-Lehman syndrome (BFLS, MIM #301900). Eur J Hum Genet 2006; 14: 1233–1237. - PubMed
-
- Zweier C, Rittinger O, Bader I et al: Females with de novo aberrations in PHF6: clinical overlap of Borjeson-Forssman-Lehmann with Coffin-Siris syndrome. Am J Med Genet C Semin Med Genet 2014; 166C: 290–301. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

