Time-resolved pathogenic gene expression analysis of the plant pathogen Xanthomonas oryzae pv. oryzae
- PMID: 27165035
- PMCID: PMC4862043
- DOI: 10.1186/s12864-016-2657-7
Time-resolved pathogenic gene expression analysis of the plant pathogen Xanthomonas oryzae pv. oryzae
Abstract
Background: Plant-pathogen interactions at early stages of infection are important to the fate of interaction. Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight, which is a devastating disease in rice. Although in vivo and in vitro systems have been developed to study rice-Xoo interactions, both systems have limitations. The resistance mechanisms in rice can be better studied by the in vivo approach, whereas the in vitro systems are suitable for pathogenicity studies on Xoo. The current in vitro system uses minimal medium to activate the pathogenic signal (expression of pathogenicity-related genes) of Xoo, but lacks rice-derived factors needed for Xoo activation. This fact emphasizes the need of developing a new in vitro system that allow for an easy control of both pathogenic activation and for the experiment itself.
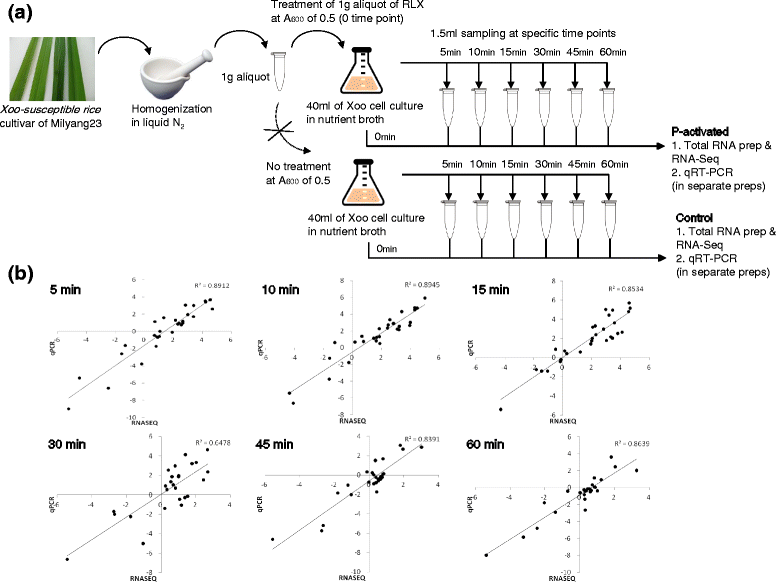
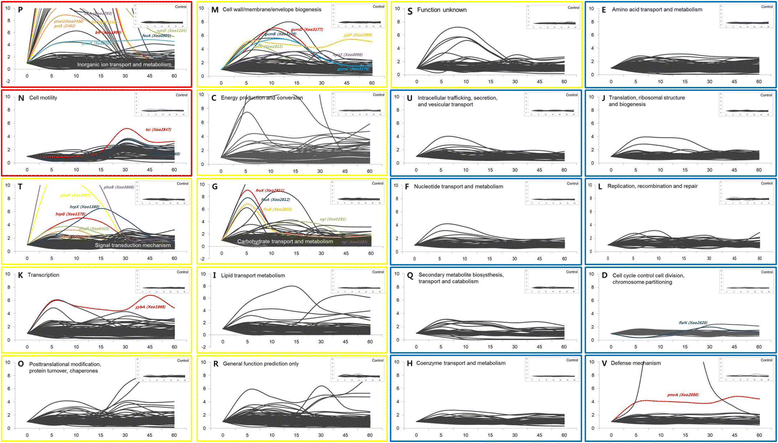
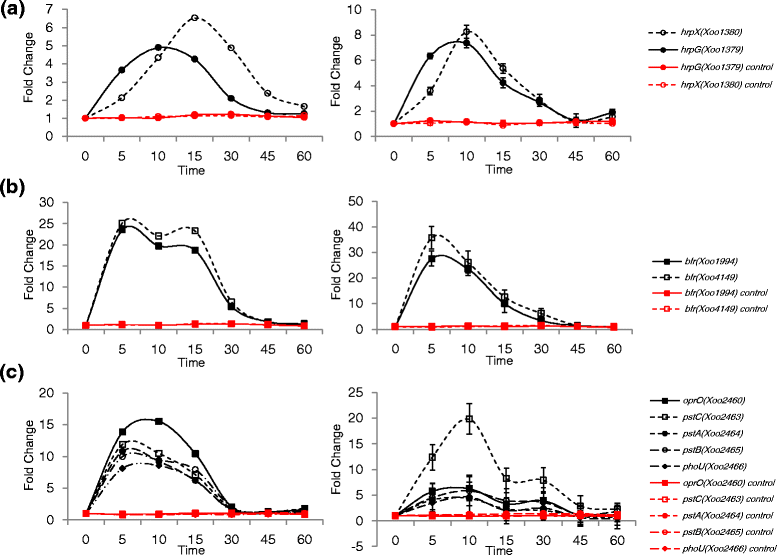
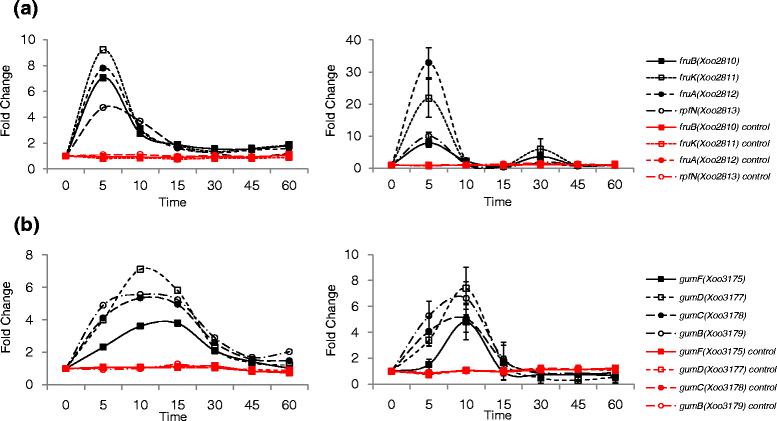
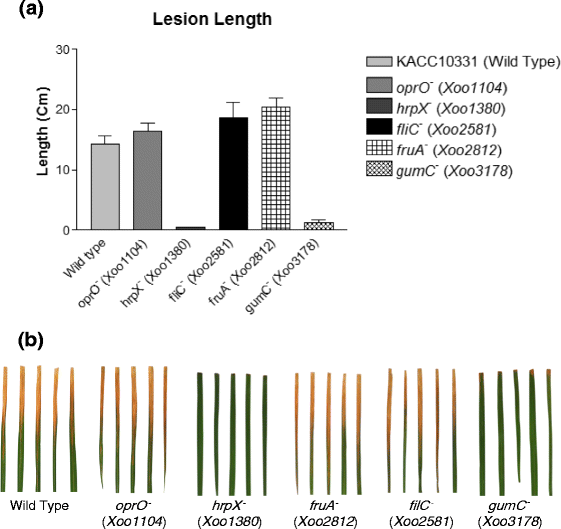
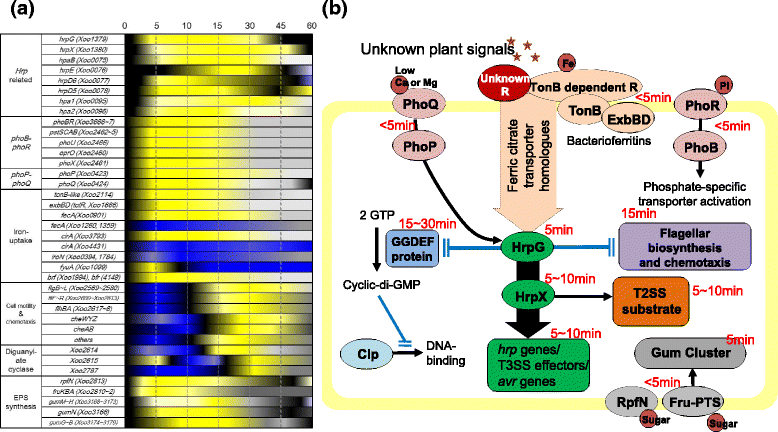
Results: We employed an in vitro system that can activate pathogenicity-related genes in Xoo using rice leaf extract (RLX) and combined the in vitro assay with RNA-Seq to analyze the time-resolved genome-wide gene expression of Xoo. RNA-Seq was performed with samples from seven different time points within 1 h post-RLX treatment and the expression of up- or downregulated genes in RNA-Seq was validated by qRT-PCR. Global analysis of gene expression and regulation revealed the most dramatic changes in functional categories of genes related to inorganic ion transport and metabolism, and cell motility. Expression of many pathogenicity-related genes was induced within 15 min upon contact with RLX. hrpG and hrpX expression reached the maximum level within 10 and 15 min, respectively. Chemotaxis and flagella biosynthesis-related genes and cyclic-di-GMP controlling genes were downregulated for 10 min and were then upregulated. Genes related to inorganic ion uptake were upregulated within 5 min. We introduced a non-linear regression fit to generate continuous time-resolved gene expression levels and tested the essentiality of the transcriptionally upregulated genes by a pathogenicity assay of lesion length using single-gene knock-out Xoo strains.
Conclusions: The in vitro system combined with RNA-Seq generated a genome-wide time-resolved pathogenic gene expression profile within 1 h of initial rice-Xoo interactions, demonstrating the expression order and interaction dependency of pathogenic genes. This combined system can be used as a novel tool to study the initial interactions between rice and Xoo during bacterial blight progression.
Keywords: Pathogenicity; Plant–pathogen interactions; RNA-Seq; Time-resolved genome-wide gene expression; Xanthomonas oryzae pv. oryzae.
Figures
References
-
- Mew TW, Alvarez AM, Leach JE, Swings J. Focus on bacterial-blight of rice. Plant Dis. 1993;77(1):5–12. doi: 10.1094/PD-77-0005. - DOI
-
- Suh JP, Jeung JU, Noh TH, Cho YC, Park SH, Park HS, Shin MS, Kim CK, Jena KK. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice. 2013;6(1):5. doi: 10.1186/1939-8433-6-5. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

