The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions
- PMID: 27165283
- PMCID: PMC4893068
- DOI: 10.1007/s00424-016-1819-4
The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions
Abstract
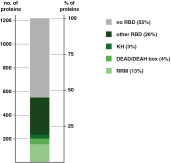
Post-transcriptional regulation of gene expression plays a critical role in almost all cellular processes. Regulation occurs mostly by RNA-binding proteins (RBPs) that recognise RNA elements and form ribonucleoproteins (RNPs) to control RNA metabolism from synthesis to decay. Recently, the repertoire of RBPs was significantly expanded owing to methodological advances such as RNA interactome capture. The newly identified RNA binders are involved in diverse biological processes and belong to a broad spectrum of protein families, many of them exhibiting enzymatic activities. This suggests the existence of an extensive crosstalk between RNA biology and other, in principle unrelated, cell functions such as intermediary metabolism. Unexpectedly, hundreds of new RBPs do not contain identifiable RNA-binding domains (RBDs), raising the question of how they interact with RNA. Despite the many functions that have been attributed to RNA, our understanding of RNPs is still mostly governed by a rather protein-centric view, leading to the idea that proteins have evolved to bind to and regulate RNA and not vice versa. However, RNPs formed by an RNA-driven interaction mechanism (RNA-determined RNPs) are abundant and offer an alternative explanation for the surprising lack of classical RBDs in many RNA-interacting proteins. Moreover, RNAs can act as scaffolds to orchestrate and organise protein networks and directly control their activity, suggesting that nucleic acids might play an important regulatory role in many cellular processes, including metabolism.
Keywords: Interactome capture; Protein-RNA interaction; RNA binding domain; RNA-binding proteins; RNA-determined RNPs; Ribonucleoproteins.
Figures
References
-
- Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Reports. 2015 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

