Olig2-Dependent Reciprocal Shift in PDGF and EGF Receptor Signaling Regulates Tumor Phenotype and Mitotic Growth in Malignant Glioma
- PMID: 27165742
- PMCID: PMC4946168
- DOI: 10.1016/j.ccell.2016.03.027
Olig2-Dependent Reciprocal Shift in PDGF and EGF Receptor Signaling Regulates Tumor Phenotype and Mitotic Growth in Malignant Glioma
Abstract
Malignant gliomas exhibit extensive heterogeneity and poor prognosis. Here we identify mitotic Olig2-expressing cells as tumor-propagating cells in proneural gliomas, elimination of which blocks tumor initiation and progression. Intriguingly, deletion of Olig2 resulted in tumors that grow, albeit at a decelerated rate. Genome occupancy and expression profiling analyses reveal that Olig2 directly activates cell-proliferation machinery to promote tumorigenesis. Olig2 deletion causes a tumor phenotypic shift from an oligodendrocyte precursor-correlated proneural toward an astroglia-associated gene expression pattern, manifest in downregulation of platelet-derived growth factor receptor-α and reciprocal upregulation of epidermal growth factor receptor (EGFR). Olig2 deletion further sensitizes glioma cells to EGFR inhibitors and extends the lifespan of animals. Thus, Olig2-orchestrated receptor signaling drives mitotic growth and regulates glioma phenotypic plasticity. Targeting Olig2 may circumvent resistance to EGFR-targeted drugs.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

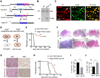
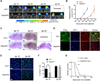
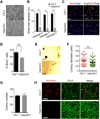

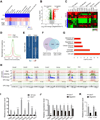
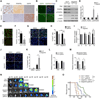
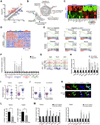
Comment in
-
Head of the Class: OLIG2 and Glioblastoma Phenotype.Cancer Cell. 2016 May 9;29(5):613-615. doi: 10.1016/j.ccell.2016.04.007. Cancer Cell. 2016. PMID: 27165737
Similar articles
-
Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas.Cell Cycle. 2017 Sep 17;16(18):1654-1660. doi: 10.1080/15384101.2017.1361062. Epub 2017 Aug 14. Cell Cycle. 2017. PMID: 28806136 Free PMC article. Review.
-
Lineage-Restricted OLIG2-RTK Signaling Governs the Molecular Subtype of Glioma Stem-like Cells.Cell Rep. 2016 Sep 13;16(11):2838-2845. doi: 10.1016/j.celrep.2016.08.040. Cell Rep. 2016. PMID: 27626655 Free PMC article.
-
Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma.Int J Cancer. 2012 Oct 1;131(7):E1078-87. doi: 10.1002/ijc.27606. Epub 2012 May 17. Int J Cancer. 2012. PMID: 22514120
-
Id2 mediates oligodendrocyte precursor cell maturation arrest and is tumorigenic in a PDGF-rich microenvironment.Cancer Res. 2014 Mar 15;74(6):1822-32. doi: 10.1158/0008-5472.CAN-13-1839. Epub 2014 Jan 14. Cancer Res. 2014. PMID: 24425046 Free PMC article.
-
Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2.Nat Rev Neurosci. 2012 Dec;13(12):819-31. doi: 10.1038/nrn3386. Nat Rev Neurosci. 2012. PMID: 23165259 Free PMC article. Review.
Cited by
-
Efficacy of osimertinib against EGFRvIII+ glioblastoma.Oncotarget. 2020 Jun 2;11(22):2074-2082. doi: 10.18632/oncotarget.27599. eCollection 2020 Jun 2. Oncotarget. 2020. PMID: 32547705 Free PMC article.
-
G-protein-coupled receptor GPR17 inhibits glioma development by increasing polycomb repressive complex 1-mediated ROS production.Cell Death Dis. 2021 Jun 12;12(6):610. doi: 10.1038/s41419-021-03897-0. Cell Death Dis. 2021. PMID: 34120140 Free PMC article.
-
Developmental origins and oncogenic pathways in malignant brain tumors.Wiley Interdiscip Rev Dev Biol. 2019 Jul;8(4):e342. doi: 10.1002/wdev.342. Epub 2019 Apr 3. Wiley Interdiscip Rev Dev Biol. 2019. PMID: 30945456 Free PMC article. Review.
-
Long noncoding RNA SNHG4: a novel target in human diseases.Cancer Cell Int. 2021 Oct 30;21(1):583. doi: 10.1186/s12935-021-02292-1. Cancer Cell Int. 2021. PMID: 34717631 Free PMC article. Review.
-
lncRNA Functional Networks in Oligodendrocytes Reveal Stage-Specific Myelination Control by an lncOL1/Suz12 Complex in the CNS.Neuron. 2017 Jan 18;93(2):362-378. doi: 10.1016/j.neuron.2016.11.044. Epub 2016 Dec 29. Neuron. 2017. PMID: 28041882 Free PMC article.
References
-
- Barrett LE, Granot Z, Coker C, Iavarone A, Hambardzumyan D, Holland EC, Nam HS, Benezra R. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer Cell. 2012;21:11–24. - PubMed
-
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

