Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma
- PMID: 27165744
- PMCID: PMC4864952
- DOI: 10.1016/j.ccell.2016.04.002
Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma
Erratum in
-
Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma.Cancer Cell. 2016 Aug 8;30(2):363. doi: 10.1016/j.ccell.2016.07.013. Epub 2016 Aug 8. Cancer Cell. 2016. PMID: 27505681 No abstract available.
Abstract
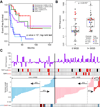
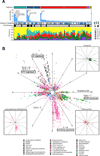
We describe a comprehensive genomic characterization of adrenocortical carcinoma (ACC). Using this dataset, we expand the catalogue of known ACC driver genes to include PRKAR1A, RPL22, TERF2, CCNE1, and NF1. Genome wide DNA copy-number analysis revealed frequent occurrence of massive DNA loss followed by whole-genome doubling (WGD), which was associated with aggressive clinical course, suggesting WGD is a hallmark of disease progression. Corroborating this hypothesis were increased TERT expression, decreased telomere length, and activation of cell-cycle programs. Integrated subtype analysis identified three ACC subtypes with distinct clinical outcome and molecular alterations which could be captured by a 68-CpG probe DNA-methylation signature, proposing a strategy for clinical stratification of patients based on molecular markers.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


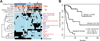


Comment in
-
Adrenal gland: The genetics of adrenocortical carcinoma revealed.Nat Rev Endocrinol. 2016 Aug;12(8):433. doi: 10.1038/nrendo.2016.89. Epub 2016 Jun 3. Nat Rev Endocrinol. 2016. PMID: 27259178 No abstract available.
References
-
- Anselmo J, Medeiros S, Carneiro V, Greene E, Levy I, Nesterova M, Lyssikatos C, Horvath A, Carney JA, Stratakis CA. A large family with Carney complex caused by the S147G PRKAR1A mutation shows a unique spectrum of disease including adrenocortical cancer. J Clin Endocrinol Metab. 2012;97:351–359. - PMC - PubMed
-
- Assie G, Letouze E, Fassnacht M, Jouinot A, Luscap W, Barreau O, Omeiri H, Rodriguez S, Perlemoine K, Rene-Corail F, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46:607–612. - PubMed
-
- Bertherat J, Groussin L, Sandrini F, Matyakhina L, Bei T, Stergiopoulos S, Papageorgiou T, Bourdeau I, Kirschner LS, Vincent-Dejean C, et al. Molecular and functional analysis of PRKAR1A and its locus (17q22–24) in sporadic adrenocortical tumors: 17q losses, somatic mutations, and protein kinase A expression and activity. Cancer Res. 2003;63:5308–5319. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- U24 CA143866/CA/NCI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U24 CA143843/CA/NCI NIH HHS/United States
- U24 CA210974/CA/NCI NIH HHS/United States
- U24 CA143858/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U24 CA143835/CA/NCI NIH HHS/United States
- U24 CA143845/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U24 CA144025/CA/NCI NIH HHS/United States
- U24 CA180951/CA/NCI NIH HHS/United States
- U24 CA143840/CA/NCI NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- U24 CA199461/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

