Integrated Genomics for Pinpointing Survival Loci within Arm-Level Somatic Copy Number Alterations
- PMID: 27165745
- PMCID: PMC4864611
- DOI: 10.1016/j.ccell.2016.03.025
Integrated Genomics for Pinpointing Survival Loci within Arm-Level Somatic Copy Number Alterations
Abstract
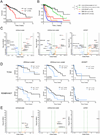
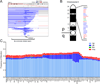
The identification of driver loci underlying arm-level somatic copy number alterations (SCNAs) in cancer has remained challenging and incomplete. Here, we assess the relative impact and present a detailed landscape of arm-level SCNAs in 10,985 patient samples across 33 cancer types from The Cancer Genome Atlas (TCGA). Furthermore, using chromosome 9p loss in lower grade glioma (LGG) as a model, we employ a unique multi-tiered genomic dissection strategy using 540 patients from three independent LGG datasets to identify genetic loci that govern tumor aggressiveness and poor survival. This comprehensive approach uncovered several 9p loss-specific prognostic markers, validated existing ones, and redefined the impact of CDKN2A loss in LGG.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




References
-
- Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer journal. 2014;20:66–72. - PubMed
-
- Beatty BG, Qi S, Pienkowska M, Herbrick JA, Scheidl T, Zhang ZM, Kola I, Scherer SW, Seth A. Chromosomal localization of phospholipase A2 activating protein, an Ets2 target gene, to 9p21. Genomics. 1999;62:529–532. - PubMed
-
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11891–11896. - PMC - PubMed
-
- Bello MJ, de Campos JM, Vaquero J, Kusak ME, Sarasa JL, Pestana A, Rey JA. Molecular and cytogenetic analysis of chromosome 9 deletions in 75 malignant gliomas. Genes, chromosomes & cancer. 1994;9:33–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

