Identification of a new gene regulatory circuit involving B cell receptor activated signaling using a combined analysis of experimental, clinical and global gene expression data
- PMID: 27166259
- PMCID: PMC5216924
- DOI: 10.18632/oncotarget.9219
Identification of a new gene regulatory circuit involving B cell receptor activated signaling using a combined analysis of experimental, clinical and global gene expression data
Abstract
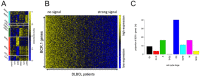
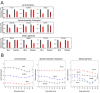
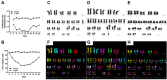
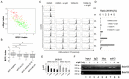
To discover new regulatory pathways in B lymphoma cells, we performed a combined analysis of experimental, clinical and global gene expression data. We identified a specific cluster of genes that was coherently expressed in primary lymphoma samples and suppressed by activation of the B cell receptor (BCR) through αIgM treatment of lymphoma cells in vitro. This gene cluster, which we called BCR.1, includes numerous cell cycle regulators. A reduced expression of BCR.1 genes after BCR activation was observed in different cell lines and also in CD10+ germinal center B cells. We found that BCR activation led to a delayed entry to and progression of mitosis and defects in metaphase. Cytogenetic changes were detected upon long-term αIgM treatment. Furthermore, an inverse correlation of BCR.1 genes with c-Myc co-regulated genes in distinct groups of lymphoma patients was observed. Finally, we showed that the BCR.1 index discriminates activated B cell-like and germinal centre B cell-like diffuse large B cell lymphoma supporting the functional relevance of this new regulatory circuit and the power of guided clustering for biomarker discovery.
Keywords: B cell receptor signaling; cell cycle delay; chromosomal aberrations; guided clustering; lymphoma.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. - PubMed
-
- Coiffier B. State-of-the-art therapeutics: diffuse large B-cell lymphoma. J Clin Oncol. 2005;23:6387–6393. - PubMed
-
- Martelli M, Ferreri AJ, Agostinelli C, Di Rocco A, Pfreundschuh M, Pileri SA. Diffuse large B-cell lymphoma. Critical reviews in oncology/hematology. 2013;87:146–171. - PubMed
-
- Kubuschok B, Held G, Pfreundschuh M. Management of diffuse large B-cell lymphoma (DLBCL) Cancer treatment and research. 2015;165:271–288. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

