Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia
- PMID: 27166522
- PMCID: PMC4922445
- DOI: 10.1080/15548627.2016.1164357
Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia
Abstract
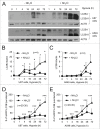
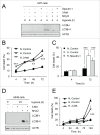
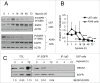
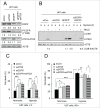
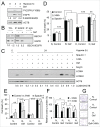
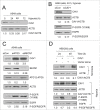
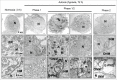
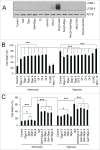
Autophagy is an intracellular lysosomal degradation pathway where its primary function is to allow cells to survive under stressful conditions. Autophagy is, however, a double-edge sword that can either promote cell survival or cell death. In cancer, hypoxic regions contribute to poor prognosis due to the ability of cancer cells to adapt to hypoxia in part through autophagy. In contrast, autophagy could contribute to hypoxia induced cell death in cancer cells. In this study, we showed that autophagy increased during hypoxia. At 4 h of hypoxia, autophagy promoted cell survival whereas, after 48 h of hypoxia, autophagy increased cell death. Furthermore, we found that the tyrosine phosphorylation of EGFR (epidermal growth factor receptor) decreased after 16 h in hypoxia. Furthermore, EGFR binding to BECN1 in hypoxia was significantly higher at 4 h compared to 72 h. Knocking down or inhibiting EGFR resulted in an increase in autophagy contributing to increased cell death under hypoxia. In contrast, when EGFR was reactivated by the addition of EGF, the level of autophagy was reduced which led to decreased cell death. Hypoxia led to autophagic degradation of the lipid raft protein CAV1 (caveolin 1) that is known to bind and activate EGFR in a ligand-independent manner during hypoxia. By knocking down CAV1, the amount of EGFR phosphorylation was decreased in hypoxia and amount of autophagy and cell death increased. This indicates that the activation of EGFR plays a critical role in the switch between cell survival and cell death induced by autophagy in hypoxia.
Keywords: Autophagy; CAV1 (caveolin-1); autosis; cell death; epidermal growth factor receptor (EGFR); hypoxia.
Figures



References
-
- Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci 2011; 124:161-70; PMID:21187343; http://dx.doi.org/10.1242/jcs.064576 - DOI - PMC - PubMed
-
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/10.1038/nature06639 - DOI - PMC - PubMed
-
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013; 368:651-62; PMID:23406030; http://dx.doi.org/10.1056/NEJMra1205406 - DOI - PubMed
-
- Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev 2014; 26:16-23; PMID:24907664; http://dx.doi.org/10.1016/j.gde.2014.04.003 - DOI - PMC - PubMed
-
- King JS. Autophagy across the eukaryotes: is S. cerevisiae the odd one out? Autophagy 2012; 8:1159-62; PMID:22722653; http://dx.doi.org/10.4161/auto.20527 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
