Meandering Down the Energy Landscape of Protein Folding: Are We There Yet?
- PMID: 27166801
- PMCID: PMC4940508
- DOI: 10.1016/j.bpj.2016.03.030
Meandering Down the Energy Landscape of Protein Folding: Are We There Yet?
Abstract
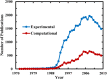
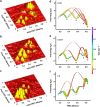
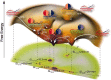
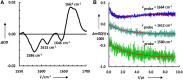
As judged by a single publication metric, the activity in the protein folding field has been declining over the past 5 years, after enjoying a decade-long growth. Does this development indicate that the field is sunsetting or is this decline only temporary? Upon surveying a small territory of its landscape, we find that the protein folding field is still quite active and many important findings have emerged from recent experimental studies. However, it is also clear that only continued development of new techniques and methods, especially those enabling dissection of the fine details and features of the protein folding energy landscape, will fuel this old field to move forward.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Levinthal C. Are there pathways for protein folding. J. Chim. Phys. Physico-Chemie Biol. 1968;65:44–45.
-
- Bryngelson J.D., Onuchic J.N., Wolynes P.G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. - PubMed
-
- Dill K.A., Chan H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. - PubMed
-
- Matouschek A., Kellis J.T., Jr., Fersht A.R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

